Journal:Molmil: A molecular viewer for the PDB and beyond
| Full article title | Molmil: A molecular viewer for the PDB and beyond |
|---|---|
| Journal | Journal of Cheminformatics |
| Author(s) | Bekker, G.J.; Nakamura, H.; Kinjo, A.R. |
| Author affiliation(s) | Osaka University |
| Primary contact | Email: gertjan dot bekker at protein dot osaka-u dot ac dot jp |
| Year published | 2016 |
| Volume and issue | 8 (1) |
| Page(s) | 42 |
| DOI | 10.1186/s13321-016-0155-1 |
| ISSN | 1758-2946 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://jcheminf.springeropen.com/articles/10.1186/s13321-016-0155-1 |
| Download | http://jcheminf.springeropen.com/track/pdf/10.1186/s13321-016-0155-1 (PDF) |
Abstract
We have developed a new platform-independent web-based molecular viewer using JavaScript and WebGL. The molecular viewer, Molmil, has been integrated into several services offered by Protein Data Bank Japan and can be easily extended with new functionality by third-party developers. Furthermore, the viewer can be used to load files in various formats from the user’s local hard drive without uploading the data to a server. Molmil is available for all platforms supporting WebGL (e.g., Windows, Linux, iOS, Android) from http://gjbekker.github.io/molmil/. The source code is available at http://github.com/gjbekker/molmil under the LGPLv3 license.
Keywords: WebGL, protein structure visualization, molecular dynamics
Background
Molecular viewers are a vital tool for our understanding of protein structures and functions. The shift from regular desktop platforms such as Windows, Mac OSX and Linux to mobile platforms such as iOS and Android in the last half-decade, however, prevents traditional online molecular viewers such as PDBj’s previously developed jV[1] and the popular Jmol[2] from running on these new platforms as these platforms do not support Java Applets. For mobile platforms a native application (i.e., an application specifically designed and optimized for each of these platforms) can be created and distributed via their respective application stores. However, with new platforms on the horizon, or already available, in addition to the already established desktop platforms, it would be a tedious and inefficient job to make a molecular viewer available on all platforms, current and future.
In the same half-decade however, web browsers have also made significant advances, particularly with their JavaScript engines. Further advances in the form of CSS3[3] and HTML5[4] have made the web an interesting platform for developers. Mobile platforms also offer these advanced web browsers, while some newer mobile platforms such as Mozilla’s Firefox OS[5] and Ubuntu Mobile[6] are oriented around HTML5 applications. Since these advanced web browsers are available on a wide range of platforms, it seems to be the perfect platform to develop a molecular viewer for.
One of the earliest attempts to switch to a Java independent molecular web viewer was done by the developers of the popular molecular viewer Jmol who developed an alternate molecular viewer named JSmol[7] to run directly within the web browser. They simply used a program known as java2script to convert Jmol’s Java code into JavaScript. JSmol however runs noticeably slower compared to Jmol for relatively small protein structures and quickly becomes unusable for larger structures. Desktop applications such as Chimera[8], VMD[9], PyMol[10] and Yasara[11] make use of GPU based hardware acceleration using OpenGL.[12] By making use of OpenGL, one can leverage the immense power of GPUs to accelerate these molecular viewers. GPUs have traditionally been designed for this purpose; i.e., to accelerate the generation of 3D images, but more recently GPUs have also been re-purposed for general purpose calculations in, for example, molecular dynamics.[13][14] So wouldn’t it be great if this power were somehow also available inside web browsers?
Enter WebGL. WebGL was developed by Khronos[15] to allow JavaScript applications running in the web browser to take advantage of OpenGL ES 2.0[16] compatible GPUs which had been specifically designed for mobile devices. While WebGL has been available for several years in Chrome[17] and Firefox[18], WebGL support was only recently added to Microsoft’s Internet Explorer and Apple’s Safari, including iOS. We have developed a new web-based molecular viewer, Molmil, which can take advantage of GPU hardware acceleration using WebGL. It runs on a wide range of platforms such as Windows, Linux, Mac OSX, Android and iOS. Finally, we have embedded Molmil within various services offered by Protein Data Bank Japan[1][19][20][21] to demonstrate its capabilities.
Implementation and capabilities
Molmil has been designed as a lightweight and full-featured viewer for the PDB. As such, Molmil can load legacy PDB flat files, PDBx/mmCIF[22] and PDBML[23] formatted files. Molmil can also load a custom format which we call PDBx/mmJSON, which is a JSON[24] version of the PDBx/mmCIF data. Other formats which Molmil supports are GRO, MOL2, MDL, CCP4[25] (for electron density maps and EM data), MyPresto’s trajectory format[14][26], Gromacs’ TRR and XTC trajectory formats[13] and our own developed MPBF polygon format which we are using for our eF-site service[1] for large structures. Users can also load these files from their local hard drive.
The PDBx/mmJSON (mmJSON for short) format was specifically developed for Molmil. In a nutshell, it is a JSON representation of the mmCIF format, made by translating the data structure based on the STAR syntax[27], into a series of associative arrays, standard arrays, integers, floats and null values. A major advantage of this format is that it can be directly interpreted by modern browsers, that is, a typed data structure is available once the JSON data has been parsed by a browser. Such is not the case for PDBML (XML) or mmCIF formats. In addition, the file size is generally smaller in the mmJSON format than in PDBML or mmCIF formats. An analysis showed that the compressed mmJSON is on average approximately 33 or 56 % smaller than a compressed mmCIF or PDBML formatted file, respectively, making it more suitable for web deployment. For large structures another derivative of mmJSON is currently under development to only include the minimal amount of information in the file required to display the backbone structure. This mmJSON variant is not yet offered as a service by PDBj, but can be previewed by using Molmil: http://gjbekker.github.io/molmil/#molmil.loadPDBlite(‘3j3q’). Implementation details regarding the parsers used to load the mmCIF data, as well as information regarding the mmJSON format and its relation with the mmCIF format is given on its Github page at http://github.com/gjbekker/cif-parsers.
Displaying large entries
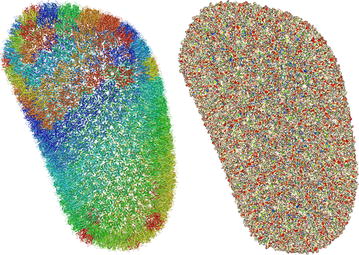
One of the design goals of Molmil was to create a molecular viewer which can produce high-quality images suitable for publications. On the other hand, it should also be able to scale to very large structures available in the Protein Data Bank (PDB) consisting of hundreds of thousands to millions of atoms. By default WebGL can only display a limited amount of polygons. However, by using an extension to WebGL (OES_element_index_uint) which is available on all modern platforms, it becomes possible to efficiently render very large or highly detailed structures. Molmil uses this extension to build high quality geometry for small and medium sized structures and dynamically scale down the quality as the size of the structure increases to gigantic proportions (such as the HIV-1 capsid; 3j3q[28], see Fig. 1). Note that to be able to load these gigantic structures, an adequate amount of memory is still required, which is often not available on smartphones, tablets and older systems. To enable high-quality lighting even when using polygon models of low detail for these gigantic structures, Molmil uses tuned Phong shading[29] which can accurately calculate the lighting even for simplistic polygon models.
|
Rendering and coloring modes
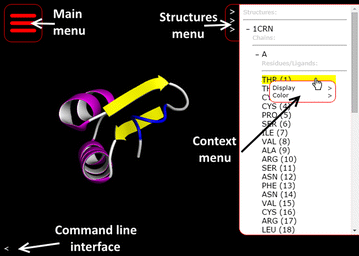
Molmil supports several rendering and coloring modes which can be selected using a point-and-click based menu. In the top-right corner of Molmil’s canvas the structures menu can be enabled (see also Fig. 2). This structures menu lists all the loaded structures in the current canvas. Via a hierarchical tree selection system chains and residues belonging to the structure can be explored. Right clicking on the structures, chains or residues displays a context menu via which users can change the display and color modes. Double clicking on the residues causes Molmil to focus and jump to the selected residue. Furthermore, right-clicking on the atoms or cartoon rendering within the canvas also enables users to interactively modify the rendering and color mode of the selected atom or the residue/chain the atom belongs to. These controls are reminiscent from Yasara[11], which uses a similar hierarchical structures menu and context menu to modify the display and color modes of the loaded structures. Molmil currently supports VDW, ball-and-stick, stick, Cα-trace, tube, cartoon, rocket and coarse surface representations. Structures can be colored by their secondary structure elements, CPK, group (blue-to-red gradient), chain, ABEGO[30] or custom color assignments. Views can be outputted as high quality PNG images, suitable for publication without requiring additional plugins. An overview of Molmil’s various rendering modes can be found in Additional file 1: Appendix I.
|
Playing animations
It is also possible to load PDB files with multiple models and play them as animations, by using the “Animation” control panel which is accessible from Molmil’s main menu (see Fig. 2). PDBj’s Promode Elastic service[20] uses this functionality. It is also possible to use this animation functionality using alternative formats to, for example, play molecular dynamics trajectories. Currently myPresto’s[26] trajectory format and Gromacs’s[13] TRR and XTC trajectory formats are supported by Molmil and support for other formats can be easily added. Although web browsers can be used to produce PNG images by Molmil, they cannot make MP4 movies. However, by using a server-side REST service it is possible to stitch the images of each frame in the trajectory together to build a movie. For obvious reasons (potential abuse) we have not made this functionality available as a public REST service, but users can obtain this application from Molmil’s Github page which they can then run on their own workstation so that their own machine will then build the movie. Using this tool, Molmil can be used to produce high quality MP4 videos of molecular simulation trajectories.
Command-line interface
Molmil also has an embedded command line which can be used to manually perform actions. The command line is capable of executing arbitrary JavaScript code and has access to Molmil’s application program interface (API), which can also be used by third party developers to embed and extend Molmil within their own web page/app. The command line can also be used to perform simple tasks such as selecting chains, residues or atoms and changing the display and coloring of them. Third-party developers can also add support for their own custom file formats, or add additional functionality. The embedded viewers on the PDBj website have been implemented in this manner. Details and examples of the Molmil API are described on Molmil’s Github page http://github.com/gjbekker/molmil/wiki. The command line can be accessed via the “<” symbol in the lower left corner of the canvas (see also Fig. 2). Finally, the commands can also be embedded within the URL for easy sharing via email, web pages and social media (see Fig. 2 for a simple example URL).
Molmil availability
Molmil has seen several major revisions since its initial release in December 2013. Molmil Beta featured a prototype WebGL viewer which was implemented directly into PDBj’s web application. Molmil 0.9 was released in September 2014 and had all the basic functionalities a molecular viewer needs such as reading various formats and supporting multiple display and color modes via a point-and-click based menu. The latest major version of Molmil, Molmil 1.0, was released in October 2015 which focused on optimizations and extending the feature set of Molmil and also added a powerful JavaScript based command line interface.
Molmil has been integrated in many of PDBj’s services[21], including: Mine PDB Explorer Asymmetric Unit viewer; Mine PDB Explorer Biological Unit viewer; EDMap viewer (for electron densities); Sequence Navigator; Structure Navigator; Promode Elastic (for viewing protein mobility analysed via Normal Mode Analysis) and Chemie (PDBj’s new chemical component service). More recently, the eF-site[1] service (for viewing the electrostatic surface of proteins) was also updated with an integrated Molmil viewer and the Biological Unit viewer was also upgraded. Furthermore, high quality PNG images of all the released PDB entries on the PDBj website have also been generated by Molmil in an automated manner. More information regarding Molmil as well as use-cases and command-line examples can be found at http://github.com/gjbekker/molmil/wiki.
Discussion
Molmil requires the browser to support WebGL. Furthermore, for optimal use, the OES_element_index_uint must also be supported. When loading very large structures such as 3j3q, there are additional requirements to the user’s software and hardware. Since Molmil requires at least six gigabytes of memory to load the structure into memory, the user’s device must be equipped with eight gigabytes of memory. Furthermore, to enable Molmil to make use of this large amount of memory, a 64-bit browser and operating system are required. Finally, due to limitations of Chrome’s JavaScript engine, even Chrome’s 64-bits version is incapable of loading 3j3q. Mozilla’s Firefox and Apple’s Safari however have no such problem with the 64-bit versions. Note that it will still take tens of seconds to several minutes depending on the web browser, the user’s hardware such as the processor and the user’s internet connection since 3j3q’s compressed data file is still about 40 megabytes large. The implementation at PDBj’s web application will automatically detect the user’s software and hardware environment and will display a link to the entry’s embedded Molmil page if the entry is compatible with the user’s system. All regular entries and most large structure entries however have no problems loading on low-end devices, as long as WebGL is supported.
Although Molmil exposes some of PyMol’s commands via Molmil’s command line, support is currently limited to a small subset of commands for selection, styling and coloring of structures. Also, although Molmil can be used to load CCP4 files for electron densities and generate coarse surfaces for proteins, high-quality surfaces such as MSMS and solvent-accessible surfaces are currently not yet supported, partly because of the high computational requirements involved in generating highly detailed surfaces.
Since Molmil’s initial release in 2013, other WebGL-based molecular viewers have also emerged. E.g., JSmol (JSmol/WebGL) has been extended with a basic WebGL based viewer and PV[31] was developed by SWISS-MODEL and is currently also being used by the RCSB-PDB. Other WebGL-based molecular viewers are listed in Additional file 1: Appendix II. Molmil however offers high-quality graphics due to using highly detailed polygon models when possible, while it can scale up to display very large structures by dynamically reducing the quality to render these large structures in combination with a finely tuned Phong shader for smooth and realistic shading even for lower detailed models. Furthermore, Molmil can also be used to easily load files from the users’ hard drive, including molecular simulation trajectories, and comes with a readily accessible command line interface, which is also programmable by embedding the commands in the URL, as shown in Fig. 2.
Conclusions
We have designed a versatile, high-performance, high-quality molecular viewer for the web. We have also deployed the molecular viewer on the PDBj web application as part of various services to aid users as well as demonstrate some of the capabilities of Molmil. The source code is available on GitHub as well as documentation on how to use and/or deploy Molmil on your own website. Questions or requests can be made either via Molmil’s GitHub page or via our contact page http://pdbj.org/contact.
Availability and requirements
Project name: Molmil
Project home page: http://github.com/gjbekker/molmil
Operating system(s): Platform independent
Programming language: JavaScript, CSS3, HTML5, WebGL, GLSL
Other requirements: WebGL
Licence: GNU LGPLv3
Restrictions for use by non-academics: None
Abbreviations
API: Application programming interface
CSS3: Cascade Style Sheets Version 3
EM: Electron microscopy
GPU: Graphics processing unit
HTML5: Hypertext Markup Language Version 5
MPBF: Molmil Polygon Binary Format
PDB: Protein Data Bank
PDBj: Protein Data Bank Japan
PNG: Portable Network Graphics
REST: Representational State Transfer
VDW: Van Der Waals
XML: extendible meta language
Declarations
Authors’ contributions
HN and GB conceived the idea. GB designed and coded the software, and led the writing of the manuscript. ARK helped write the manuscript and conceived the name of the program. HN and ARK contributed to the further development of the idea and in giving suggestions for improvements to the program. All authors read and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Open access
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
Additional files
- Additional file 1. Appendices; Example renderings using Molmil & Overview of alternative WebGL based molecular viewers: 13321_2016_155_MOESM1_ESM.pdf
References
- ↑ 1.0 1.1 1.2 1.3 Kinoshita, K.; Nakamura, H. (2003). "eF-site and PDBjViewer: database and viewer for protein functional sites". Bioinformatics 20 (8): 1329-1330. doi:10.1093/bioinformatics/bth073. PMID 14871866.
- ↑ Hanson, R.M. (2010). "Jmol - A paradigm shift in crystallographic visualization". Journal of Applied Crystallography 43 (5): 1250-1260. doi:10.1107/S0021889810030256.
- ↑ W3C Working Group (13 October 2015). "CSS Snapshot 2015". World Wide Web Consortium. https://www.w3.org/TR/CSS/. Retrieved 14 March 2016.
- ↑ W3C Working Group (28 October 2014). "HTML5: A vocabulary and associated APIs for HTML and XHTML". World Wide Web Consortium. https://www.w3.org/TR/html5/. Retrieved 14 March 2016.
- ↑ "Firefox OS". Mozilla. https://www.mozilla.org/en-US/firefox/os/. Retrieved 14 March 2016.
- ↑ "Ubuntu for Phones". Canonical Ltd. http://www.ubuntu.com/phone. Retrieved 14 March 2016.
- ↑ Hanson, R.M.; Prilusky, J.; Renjian, Z. et al. (2013). "JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia". Israel Journal of Chemistry 53 (3-4): 207–216. doi:10.1002/ijch.201300024.
- ↑ Pettersen, E.F.; Goddard, T.D.; Huang, C.C. et al. (2004). "UCSF Chimera—A visualization system for exploratory research and analysis". Journal of Computational Chemistry 25 (13): 1605–1612. doi:10.1002/jcc.20084. PMID 15264254.
- ↑ Humphrey, W.; Dalke, A.; Schulten, K. (1996). "VMD: Visual molecular dynamics". Journal of Molecular Graphics 14 (1): 33–38. doi:10.1016/0263-7855(96)00018-5. PMID 8744570.
- ↑ Schrödinger, L. (2010). "The PyMOL molecular graphics system, version 1.3r1". https://www.pymol.org/.
- ↑ 11.0 11.1 Krieger, E.; Vriend, G. (2014). "YASARA View - Molecular graphics for all devices, from smartphones to workstations". Bioinformatics 30 (20): 2981-2. doi:10.1093/bioinformatics/btu426. PMC PMC4184264. PMID 24996895. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4184264.
- ↑ "OpenGL — The industry standard for high performance graphics". Khronos Group. https://www.opengl.org/. Retrieved 14 March 2016.
- ↑ 13.0 13.1 13.2 Abraham, M.J.; Murtola, T.; Schulz, R. et al. (2015). "GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers". SoftwareX 1–2: 19–25. doi:10.1016/j.softx.2015.06.001.
- ↑ 14.0 14.1 Mashimo, T.; Fukunishi, Y.; Kamiya, N. et al. (2013). "Molecular dynamics simulations accelerated by GPU for biological macromolecules with a non-Ewald scheme for electrostatic interactions". Journal of Chemical Theory and Computation 9 (12): 5599–5609. doi:10.1021/ct400342e.
- ↑ "OpenGL ES 2.0 for the Web". Khronos Group. https://www.khronos.org/webgl/. Retrieved 14 December 2015.
- ↑ "OpenGL ES 2.x: The Standard for Embedded Accelerated 3D Graphics". Khronos Group. https://www.khronos.org/opengles/2_X/. Retrieved 14 March 2016.
- ↑ "Chrome Web Browser". Google. https://www.google.com/chrome/. Retrieved 14 March 2016.
- ↑ "Firefox". Mozilla. http://www.mozilla.org/en-US/firefox/new/. Retrieved 14 March 2016.
- ↑ "Protein Data Bank Japan". Protein Data Bank Japan. http://pdbj.org/. Retrieved 14 March 2016.
- ↑ 20.0 20.1 Wako, H.; Endo, S. (2013). "Normal mode analysis based on an elastic network model for biomolecules in the Protein Data Bank, which uses dihedral angles as independent variables". Computational Biology and Chemistry 44: 22-30. doi:10.1016/j.compbiolchem.2013.02.006. PMID 23537565.
- ↑ 21.0 21.1 Kinjo, A.R.; Suzuki, H.; Yamashita, R. et al. (2012). "Protein Data Bank Japan (PDBj): Maintaining a structural data archive and resource description framework format". Nucleic Acids Research 40 (D1): D453-60. doi:10.1093/nar/gkr811. PMC PMC3245181. PMID 21976737. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3245181.
- ↑ Fitzgerald, P.M.D.; Westbrook, J.D.; Bourne, P.E. et al. (2006). "Macromolecular dictionary (mmCIF)". In Hall, S.R.; McMahon, B.. International Tables for Crystallography. pp. 295–443. doi:10.1107/97809553602060000745. http://dx.doi.org/10.1107/97809553602060000745.
- ↑ Westbrook, J.; Ito, N.; Nahamura, H. et al. (2005). "PDBML: The representation of archival macromolecular structure data in XML". Bioinformatics 21 (7): 988-92. doi:10.1093/bioinformatics/bti082. PMID 15509603.
- ↑ "Introducing JSON". http://www.json.org/. Retrieved 14 March 2016.
- ↑ Winn, M.D.; Ballard, C.C.; Cowtan, K.D. et al. (2011). "Overview of the CCP4 suite and current developments". Acta Crystallographica Section D D67 (Part 4): 235-242. doi:10.1107/S0907444910045749.
- ↑ 26.0 26.1 Morikami, K.; Nakai, T.; Kidera, A. et al. (1992). "Presto (protein engineering simulator): A vectorized molecular mechanics program for biopolymers". Computers & Chemistry 16 (3): 243-248. doi:10.1016/0097-8485(92)80010-W.
- ↑ Hall, S.R. (1991). "The STAR file: A new format for electronic data transfer and archiving". Journal of Chemical Information and Modeling 31 (2): 326–333. doi:10.1021/ci00002a020.
- ↑ Zhao, G.; Perilla, J.R.; Yufenyuy, E.L. et al. (2013). "Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics". Nature 497 (7451): 643-6. doi:10.1038/nature12162. PMC PMC3729984. PMID 23719463. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3729984.
- ↑ Phong, B.T. (1975). "Illumination for computer generated pictures". Communications of the ACM 18 (6): 311–317. doi:10.1145/360825.360839.
- ↑ Lin, Y.R.; Koga, N.; Tatsumi-Koga, R. et al. (2015). "Control over overall shape and size in de novo designed proteins". Proceedings of the National Academy of Sciences of the United States of Americs 112 (40): E5478-85. doi:10.1073/pnas.1509508112. PMC PMC4603489. PMID 26396255. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4603489.
- ↑ Biasini, M. (2015). "PV". GitHub. https://github.com/biasmv/pv.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.