Difference between revisions of "Journal:AI meets exascale computing: Advancing cancer research with large-scale high-performance computing"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 59: | Line 59: | ||
As omics data continues to accumulate, computational models integrating multimodal data sources become possible. Multimodal deep learning<ref name="SunAMulti19">{{cite journal |title=A Multimodal Deep Neural Network for Human Breast Cancer Prognosis Prediction by Integrating Multi-Dimensional Data |journal=IEEE/ACM Transactions on Computational Biology and Bioinformatics |author=Sun, D.; Wang, M.; Li, A. |volume=16 |issue=3 |pages=841–50 |year=2019 |doi=10.1109/TCBB.2018.2806438}}</ref> aims to enhance learned features for one task by learning features over multiple modalities. Early Pilot One work<ref name="XiaPredict18">{{cite journal |title=Predicting tumor cell line response to drug pairs with deep learning |journal=BMC Bioinformatics |author=Xia, F.; Shukla, M.; Brettin, T. et al. |volume=19 |issue=Suppl. 18 |page=486 |year=2018 |doi=10.1186/s12859-018-2509-3 |pmid=30577754 |pmc=PMC6302446}}</ref> measured performance of multi-modal deep neural network drug pair response models with five-fold cross validation. Using the NCI-ALMANAC<ref name="HolbeckTheNat17">{{cite journal |title=The National Cancer Institute ALMANAC: A Comprehensive Screening Resource for the Detection of Anticancer Drug Pairs with Enhanced Therapeutic Activity |journal=Cancer Research |author=Holbeck, S.L.; Camalier, R.; Crowell, J.A. et al. |volume=77 |issue=13 |pages=3564-3576 |year=2017 |doi=10.1158/0008-5472.CAN-17-0489 |pmid=28446463 |pmc=PMC5499996}}</ref> data, best model performance was demonstrated when gene expression, microRNA, proteome, and Dragon7 drug descriptors<ref name="KodeDragon19">{{cite web |url=https://chm.kode-solutions.net/products_dragon.php |title=Dragon |publisher=Kode Chemoinformatics srl |date=2019 |accessdate=30 April 2019}}</ref> were combined obtaining an R-squared value of 0.944, which indicates that over 94% of the variation in tumor response is explained by the variation among the contributing gene expression, micro RNA expression, [[proteomics]], and drug property data. | As omics data continues to accumulate, computational models integrating multimodal data sources become possible. Multimodal deep learning<ref name="SunAMulti19">{{cite journal |title=A Multimodal Deep Neural Network for Human Breast Cancer Prognosis Prediction by Integrating Multi-Dimensional Data |journal=IEEE/ACM Transactions on Computational Biology and Bioinformatics |author=Sun, D.; Wang, M.; Li, A. |volume=16 |issue=3 |pages=841–50 |year=2019 |doi=10.1109/TCBB.2018.2806438}}</ref> aims to enhance learned features for one task by learning features over multiple modalities. Early Pilot One work<ref name="XiaPredict18">{{cite journal |title=Predicting tumor cell line response to drug pairs with deep learning |journal=BMC Bioinformatics |author=Xia, F.; Shukla, M.; Brettin, T. et al. |volume=19 |issue=Suppl. 18 |page=486 |year=2018 |doi=10.1186/s12859-018-2509-3 |pmid=30577754 |pmc=PMC6302446}}</ref> measured performance of multi-modal deep neural network drug pair response models with five-fold cross validation. Using the NCI-ALMANAC<ref name="HolbeckTheNat17">{{cite journal |title=The National Cancer Institute ALMANAC: A Comprehensive Screening Resource for the Detection of Anticancer Drug Pairs with Enhanced Therapeutic Activity |journal=Cancer Research |author=Holbeck, S.L.; Camalier, R.; Crowell, J.A. et al. |volume=77 |issue=13 |pages=3564-3576 |year=2017 |doi=10.1158/0008-5472.CAN-17-0489 |pmid=28446463 |pmc=PMC5499996}}</ref> data, best model performance was demonstrated when gene expression, microRNA, proteome, and Dragon7 drug descriptors<ref name="KodeDragon19">{{cite web |url=https://chm.kode-solutions.net/products_dragon.php |title=Dragon |publisher=Kode Chemoinformatics srl |date=2019 |accessdate=30 April 2019}}</ref> were combined obtaining an R-squared value of 0.944, which indicates that over 94% of the variation in tumor response is explained by the variation among the contributing gene expression, micro RNA expression, [[proteomics]], and drug property data. | ||
Mechanistically informed feature selection is an alternative approach that has the potential to increase predictive model performance. The LINCS landmark genes<ref name="SubramanianANext17">{{cite journal |title=A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles |journal=Cell |author=Subramanian, A.; Narayan, R.; Corsello, S.M. et al. |volume=171 |issue=6 |pages=P1437-1452.E17 |year=2017 |doi=10.1016/j.cell.2017.10.049 |pmid=29195078 |pmc=PMC5990023}}</ref> for example have been used to train deep learning models to predict gene expression of non-landmark genes<ref name="ChenGene16">{{cite journal |title=Gene expression inference with deep learning |journal=Bioinformatics |author=Chen, Y.; Li, Y.; Narayan, R. et al. |volume=32 |issue=12 |pages=1832-9 |year=2016 |doi=10.1093/bioinformatics/btw074 |pmid=26873929 |pmc=PMC4908320}}</ref> and to classify drug-target interactions.<ref name="XieDeep18">{{cite journal |title=Deep learning-based transcriptome data classification for drug-target interaction prediction |journal=BMC Genomics |author=Xie, L.; He, S.; Song, X. et al. |volume=19 |issue=Suppl. 7 |page=667 |year=2018 |doi=10.1186/s12864-018-5031-0 |pmid=30255785 |pmc=PMC6156897}}</ref> Ongoing work in Pilot One is exploring the impact on prediction using gene sets like that of the LINCS landmark genes and other mechanistically defined gene sets. The potential of employing mechanistically informed feature selection extends beyond improving prediction accuracy, to the realm of building models on the basis of existing biological knowledge. | |||
==References== | ==References== | ||
Revision as of 16:13, 19 November 2019
| Full article title | AI meets exascale computing: Advancing cancer research with large-scale high-performance computing |
|---|---|
| Journal | Frontiers in Oncology |
| Author(s) |
Bhattacharya, Tanmoy; Brettin, Thomas; Doroshow, James H.; Evrard, Yvonne A.; Greenspan, Emily J.; Gryshuk, Amy L.; Hoang, Thuc T.; Vea Lauzon, Carolyn, B.; Nissley, Dwight; Penberthy, Lynne; Stahlberg, Eric; Stevens, Rick; Streitz, Fred; Tourassi, Georgia; Xia, Fangfang; Zaki, George |
| Author affiliation(s) |
Los Alamos National Laboratory, Argonne National Laboratory, National Cancer Institute, Frederick National Laboratory for Cancer Research, Lawrence Livermore National Laboratory, National Nuclear Security Administration, U.S. Department of Energy Office of Science, University of Chicago, Oak Ridge National Laboratory |
| Primary contact | Email: george dot zaki at nih dot gov |
| Editors | Meerzaman, Daoud |
| Year published | 2019 |
| Volume and issue | 9 |
| Page(s) | 984 |
| DOI | 10.3389/fonc.2019.00984 |
| ISSN | 2234-943X |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.frontiersin.org/articles/10.3389/fonc.2019.00984/full |
| Download | https://www.frontiersin.org/articles/10.3389/fonc.2019.00984/pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
The application of data science in cancer research has been boosted by major advances in three primary areas: (1) data: diversity, amount, and availability of biomedical data; (2) advances in artificial intelligence (AI) and machine learning (ML) algorithms that enable learning from complex, large-scale data; and (3) advances in computer architectures allowing unprecedented acceleration of simulation and machine learning algorithms. These advances help build in silico ML models that can provide transformative insights from data, including molecular dynamics simulations, next-generation sequencing, omics, imaging, and unstructured clinical text documents. Unique challenges persist, however, in building ML models related to cancer, including: (1) access, sharing, labeling, and integration of multimodal and multi-institutional data across different cancer types; (2) developing AI models for cancer research capable of scaling on next-generation high-performance computers; and (3) assessing robustness and reliability in the AI models. In this paper, we review the National Cancer Institute (NCI) -Department of Energy (DOE) collaboration, the Joint Design of Advanced Computing Solutions for Cancer (JDACS4C), a multi-institution collaborative effort focused on advancing computing and data technologies to accelerate cancer research on the molecular, cellular, and population levels. This collaboration integrates various types of generated data, pre-exascale compute resources, and advances in ML models to increase understanding of basic cancer biology, identify promising new treatment options, predict outcomes, and, eventually, prescribe specialized treatments for patients with cancer.
Keywords: cancer research, high-performance computing, artificial intelligence, deep learning, natural language processing, multi-scale modeling, precision medicine, uncertainty quantification
Introduction
Predictive computational models for patients with cancer can in the future support prevention and treatment decisions by informing choices to achieve the best possible clinical outcome. Toward this vision, in 2015, the national Precision Medicine Initiative (PMI)[1] was announced, motivating efforts to target and advance precision oncology, including looking ahead to the scientific, data, and computational capabilities needed to advance this vision. At the same time, the horizon of computing was changing in the life sciences, as the capabilities and transformations enabled by exascale computing were coming into focus, driven by the accelerated growth in data volumes and anticipated new sources of information catalyzed by new technologies and initiatives such as PMI.
The National Strategic Computing Initiative (NSCI) in 2015 named the Department of Energy (DOE) as a lead agency for “advanced simulation through a capable exascale computing program” and the National Institutes of Health (NIH) as one of the deployment agencies to participate “in the co-design process to integrate the special requirements of their respective missions.” This interagency coordination structure opened the avenue for a tight collaboration between the NCI and the DOE. With shared aims to advance cancer research while shaping the future for exascale computing, the NCI and DOE established the Joint Design of Advanced Computing Solutions for Cancer (JDACS4C) in June of 2016 through a five-year memorandum of understanding with three co-designed pilot efforts to address both national priorities. The high-level goals of these three pilots were to push the frontiers of computing technologies in specific areas of cancer research:
- at the cellular level: advance the capabilities of patient-derived pre-clinical models to identify new treatments;
- at the molecular level: further understand the basic biology of undruggable targets; and
- at the population level: gain critical insights on the drivers of population cancer outcomes.
The pilots would also develop new uncertainty quantification (UQ) methods to evaluate confidence in the AI model predictions.
Using co-design principles, each of the pilots in the JDACS4C collaboration is based on—and driven by—team science, which is the hallmark of the collaboration's success. Enabled by deep learning, Pilot One (cellular-level) combines data in innovative ways to develop computationally predictive models for tumor response to novel therapeutic agents. Pilot Two (molecular-level) combines experimental data, simulation, and AI to provide new windows to understand and explore the biology of cancers related to the Ras superfamily of proteins. Pilot Three (population-level) uses AI and clinical information at unprecedented scales to enable precision cancer surveillance to transform cancer care.
AI and large-scale computing to predict tumor treatment response
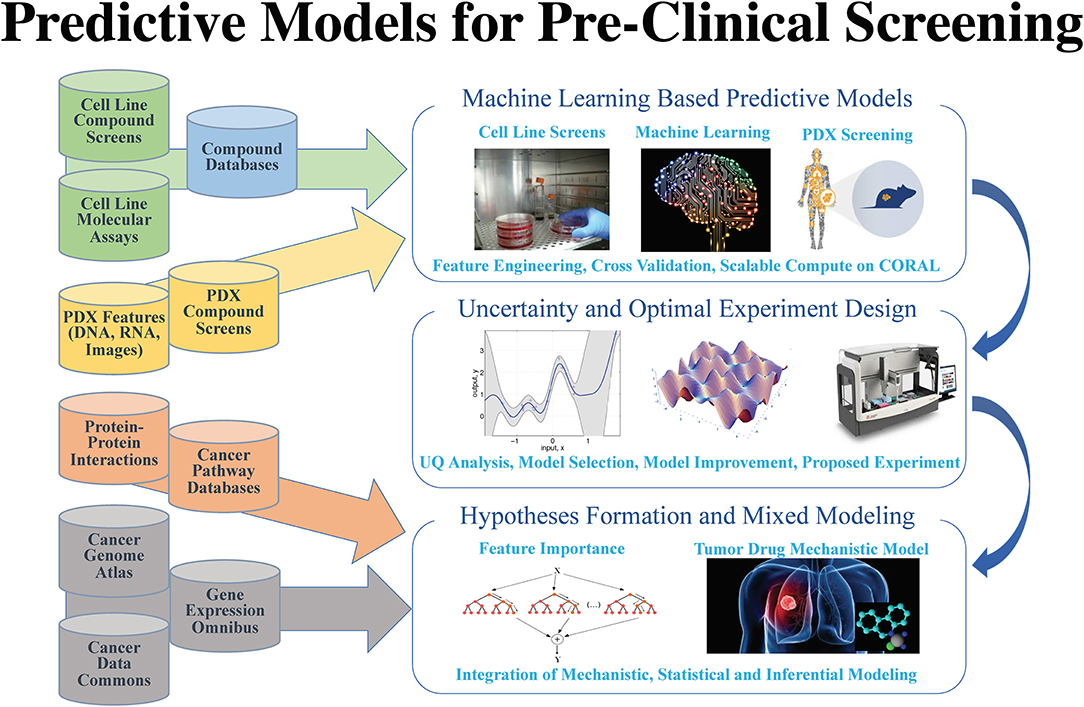
After years of efforts within the research and pharmaceutical sectors, many patients with cancer still do not respond to standard-of-care treatments, and emergence of therapy resistance is common. Efforts in precision medicine may someday change this by using a targeted therapeutics approach, individually tailored to each patient based on predictive models that use molecular and drug signatures. The Predictive Modeling for Pre-Clinical Screening Pilot (Pilot One) aims to develop predictive capabilities of drug response in pre-clinical models of cancer to improve and expedite the selection and development of new targeted therapies for patients with cancer. Highlights of the work done in Pilot One are shown in Figure 1.
|
As omics data continues to accumulate, computational models integrating multimodal data sources become possible. Multimodal deep learning[2] aims to enhance learned features for one task by learning features over multiple modalities. Early Pilot One work[3] measured performance of multi-modal deep neural network drug pair response models with five-fold cross validation. Using the NCI-ALMANAC[4] data, best model performance was demonstrated when gene expression, microRNA, proteome, and Dragon7 drug descriptors[5] were combined obtaining an R-squared value of 0.944, which indicates that over 94% of the variation in tumor response is explained by the variation among the contributing gene expression, micro RNA expression, proteomics, and drug property data.
Mechanistically informed feature selection is an alternative approach that has the potential to increase predictive model performance. The LINCS landmark genes[6] for example have been used to train deep learning models to predict gene expression of non-landmark genes[7] and to classify drug-target interactions.[8] Ongoing work in Pilot One is exploring the impact on prediction using gene sets like that of the LINCS landmark genes and other mechanistically defined gene sets. The potential of employing mechanistically informed feature selection extends beyond improving prediction accuracy, to the realm of building models on the basis of existing biological knowledge.
References
- ↑ "What is the Precision Medicine Iniative?". Genetics Home Reference. National Institutes of Health. 2019. https://ghr.nlm.nih.gov/primer/precisionmedicine/initiative. Retrieved 20 September 2019.
- ↑ Sun, D.; Wang, M.; Li, A. (2019). "A Multimodal Deep Neural Network for Human Breast Cancer Prognosis Prediction by Integrating Multi-Dimensional Data". IEEE/ACM Transactions on Computational Biology and Bioinformatics 16 (3): 841–50. doi:10.1109/TCBB.2018.2806438.
- ↑ Xia, F.; Shukla, M.; Brettin, T. et al. (2018). "Predicting tumor cell line response to drug pairs with deep learning". BMC Bioinformatics 19 (Suppl. 18): 486. doi:10.1186/s12859-018-2509-3. PMC PMC6302446. PMID 30577754. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6302446.
- ↑ Holbeck, S.L.; Camalier, R.; Crowell, J.A. et al. (2017). "The National Cancer Institute ALMANAC: A Comprehensive Screening Resource for the Detection of Anticancer Drug Pairs with Enhanced Therapeutic Activity". Cancer Research 77 (13): 3564-3576. doi:10.1158/0008-5472.CAN-17-0489. PMC PMC5499996. PMID 28446463. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5499996.
- ↑ "Dragon". Kode Chemoinformatics srl. 2019. https://chm.kode-solutions.net/products_dragon.php. Retrieved 30 April 2019.
- ↑ Subramanian, A.; Narayan, R.; Corsello, S.M. et al. (2017). "A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles". Cell 171 (6): P1437-1452.E17. doi:10.1016/j.cell.2017.10.049. PMC PMC5990023. PMID 29195078. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5990023.
- ↑ Chen, Y.; Li, Y.; Narayan, R. et al. (2016). "Gene expression inference with deep learning". Bioinformatics 32 (12): 1832-9. doi:10.1093/bioinformatics/btw074. PMC PMC4908320. PMID 26873929. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4908320.
- ↑ Xie, L.; He, S.; Song, X. et al. (2018). "Deep learning-based transcriptome data classification for drug-target interaction prediction". BMC Genomics 19 (Suppl. 7): 667. doi:10.1186/s12864-018-5031-0. PMC PMC6156897. PMID 30255785. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6156897.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.