Journal:Arkheia: Data management and communication for open computational neuroscience
| Full article title | Arkheia: Data management and communication for open computational neuroscience |
|---|---|
| Journal | Frontiers in Neuroinformatics |
| Author(s) | Antolik, Ján; Davison, Andrew P. |
| Author affiliation(s) | Institut de la Vision, Centre National de la Recherche Scientifique |
| Primary contact | Email: antolikjan at gmail dot com |
| Editors | Valdes-Sosa, Pedro Antonio |
| Year published | 2018 |
| Volume and issue | 12 |
| Page(s) | 6 |
| DOI | 10.3389/fninf.2018.00006 |
| ISSN | 1662-5196 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.frontiersin.org/articles/10.3389/fninf.2018.00006/full |
| Download | https://www.frontiersin.org/articles/10.3389/fninf.2018.00006/pdf (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Two trends have been unfolding in computational neuroscience during the last decade. First, focus has shifted to increasingly complex and heterogeneous neural network models, with a concomitant increase in the level of collaboration within the field (whether direct or in the form of building on top of existing tools and results). Second, general trends in science have shifted toward more open communication, both internally, with other potential scientific collaborators, and externally, with the wider public. This multi-faceted development toward more integrative approaches and more intense communication within and outside of the field poses major new challenges for modelers, as currently there is a severe lack of tools to help with automatic communication and sharing of all aspects of a simulation workflow to the rest of the community. To address this important gap in the current computational modeling software infrastructure, here we introduce Arkheia, a web-based open science platform for computational models in systems neuroscience. It provides an automatic, interactive, graphical presentation of simulation results, experimental protocols, and interactive exploration of parameter searches in a browser-based application. Arkheia is focused on the automatic presentation of these resources with minimal manual input from users. Arkheia is written in a modular fashion, with a focus on future development of the platform. The platform is designed in an open manner, with a clearly defined and separated application programming interface (API) for database access, so that any project can write its own back-end, translating its data into the Arkheia database format. Arkheia is not a centralized platform, but it allows any user (or group of users) to set up their own repository, either for public access by the general population, or locally for internal use. Overall, Arkheia provides users with an automatic means to communicate information about not only their models but also individual simulation results and the entire experimental context in an approachable, graphical manner, thus facilitating the user's ability to collaborate in the field and outreach to a wider audience.
Keywords: computational modeling, workflow, publish, neuroscience, tool
Introduction
For most of its history, computational neuroscience has focused on relatively homogeneous models, targeting one or at most a handful of features of neural processing at a time. Such a classical reductionist approach is starting to be supplemented by more integrative strategies that utilize increasingly complex and heterogeneous neural network models in order to explain within a single model instance an increasingly broad range of neural phenomena.[1][2][3][4][5][6] Even though the classical reductionist approach will remain important, an integrative research program seems unavoidable if we are to understand a complex dynamical system such as the cortex (or the entire brain), whose computational power is underlined by the dynamical interplay of all its anatomical and functional constituents, rather than just their simple aggregation. Given its sheer scope and complexity, such an integrative research program is unlikely to succeed if implemented by individual scientists or even individual teams. Rather, a systematic incremental strategy relying on cooperation within the entire field will be required, whereupon new models build directly on previous work, and all models are extensively validated against biological data and compared against previous models based on an increasingly exhaustive set of measures. These trends herald the shift of focus from model creation and simulation to model analysis and testing.
At the same time, this increasing need for collaboration within computational neuroscience is accompanied by a more general trend in science toward more open communication, both internally, with other potential scientific collaborators, and externally, with the wider public. Many examples have by now shown the value of such open science approaches[7][8] to promote one's research and find new collaborations. Engagement of a non-academic enthusiast audience via open-science platforms can not only improve the public outreach of one's research program, but also contribute to the core scientific development. However, the effectiveness of such an opening up of one's research is critically dependent on the ease with which outsiders can engage with the exposed resources, which in turn critically depends on the quality of the (software) infrastructure used to serve said resources.
This multi-faceted development toward more integrative approaches and intensifying communication within and outside the field poses major new challenges for the software infrastructure available to computational neuroscientists. The set of tools involved in a typical modeler's workflow is expanding concurrently with growing complexity in the metadata flowing between them. Meanwhile the requirements for their efficient interfacing with the outside world (whether in the form of human users or other software tools) is growing. This growing complexity of the tasks involved in the typical modeler's workflow is putting strain on researchers, who are required to manage increasingly more complex software infrastructure while spending a substantial portion of their work time either writing ad-hoc software solutions to cover poorly supported aspects of the workflow or handling them manually. This situation is clearly less than ideal, slowing down the pace of research while introducing errors and hindering its reproducibility.
The last four decades have seen numerous additions to the ecosystem of computational neuroscience tools, including efficient, well tested, and highly usable simulators such as Neuron[9], NEST[10], Brian[11], NENGO[12], and others; data management and parameter exploration tools such as PyNN[13], Neo[14], Lancet[15], Pypet[16], and others[17][18]; neural data analysis toolkits SpikeViewer[19], HRLAnalysis[20], NeuroTools (http://neuralensemble.org/NeuroTools), and Elephant (http://neuralensemble.org/elephant); and integrated workflow and simulation environments such as VirtualBrain[21], psychopy_ext[22], or Mozaik[23]. Despite this rapid progress, the interfacing between the tools and communication with third parties (whether users or tools) remains limited, hindering the future development of integrative collaborative approaches in computational systems neuroscience. We identify the following aspects of the modeling workflow, all with implications for communication and interfacing, that are currently poorly supported and are key to resolving the outlined limitations of the present infrastructure:
1. Higher-level, flexible, modular model specification standards allowing for transparent and efficient communication and reuse of model components
2. Exhaustive, explicitly formalized annotation of data generated during model simulation allowing for deep automatic introspection of the raw neural data in subsequent processing steps (i.e., 3 and 4)
3. Explicit formalization of experimental protocols and neural data analysis allowing for (a) automatic testing and comparison of the models, (b) their efficient communication and reuse, and (c) deep introspection of the results
4. Tools that can utilize 1, 2, and 3 to automatically communicate and serve all aspect of the modeler's workflow to the rest of the community and public to facilitate collaboration and outreach
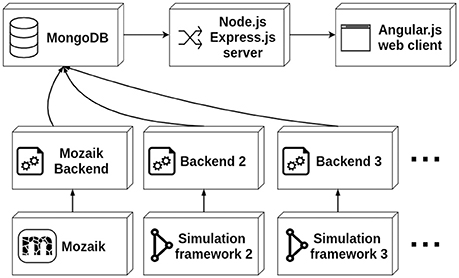
Recently we have made advances in addressing some aspect of points 1, 2, and 3 with the release of the Mozaik toolkit[23], which allows us to start approaching limitation 4, by introducing Arkheia (downloadable from https://github.com/antolikjan/Arkheia and demoed at http://arkheia.org/). Arkheia is a web-based platform for data management and communication of computational modeling outcomes in systems neuroscience. It provides an automatic, interactive graphical presentation of simulation results, the experimental protocols used, and interactive exploration of parameter searches, via a web browser-based application. Arkheia is focused on automatic serving of these resources with minimal (virtually no) manual input from users. Arkheia is written in modular fashion with a focus on future development of the platform. It follows the standard database-server-client design and is based around modern widely adopted web-based technologies (MongoDB for the database, Node.js and Express.js for the server, and AngularJS for the client). Currently, Arkheia is shipped only with a Mozaik backend, as at present this is the only published framework providing sufficient introspection of simulated neural data that can be automatically harvested for the presentation in Arkheia. The platform is, however, designed in an open manner, with a clearly defined and separated API for database access, so that any project can write its own back-end, translating its data into the Arkheia database format. This both allows any current private ad-hoc project to use Arkheia as its graphical bookkeeping and publishing front-end, as well as ensuring Arkheia can be used with any future workflow tools that may be developed. Arkheia does not currently offer an internally implemented fine-grain access control and is not meant to be used as a centralized platform. Rather, it allows any user (or group of users) to set up a separate repository, either publicly for access by the general population, as well as locally for internal use.
Overall, Arkheia provides users with an automatic means to communicate not only their models but also individual simulation results and the entire experimental context in an approachable graphical manner to a wider audience. As such, Arkheia addresses some of the limitations of the present computational neuroscience infrastructure in managing and communicating results, thus facilitating the user's ability to collaborate in the field and outreach to a wider audience.
Comparison to other tools
Several recent software projects have overlapping goals with Arkheia. Lancet[15] and Pypet[16] are Python simulation workflow libraries that provide users with a means to organize and automate their numerical simulation workflow, automate exploration of model parameter space, and manage the resulting data. Similarly to Arkheia, they provide users with structured access to the data produced in the simulations, enriched with a limited set of metadata tracked during the simulation workflow, mostly comprising the parametric configuration of the simulated models. Arkheia does not provide direct handling of the simulation workflow or the exploration parameter space. These are instead expected to be performed by the source of the data handled by Arkheia, and the Mozaik toolkit for which the data-import back-end is currently provided offers both these services. Crucially, the Lancet and Pypet toolkits do not provide a graphical interactive representation of the data to the user, which is the primary goal of Arkheia. Furthermore, Lancet and Pypet are agnostic as to the nature of the simulations they handle, which makes them more general, but at the cost of explicitly exposing only a very limited set of information about the simulations to the user. In contrast, Arkheia is focused exclusively on neural simulations, allowing it to provide the user with much richer and deeper introspection of information about the simulations held in the repository and to do it via a clear and convenient web-based graphical user interface.
Probably the most similar tool currently available to the computational neuroscience community is the Open Source Brain (OSB) (http://www.opensourcebrain.org/) project, a web-based open science collaborative platform aspiring to become the go-to repository for neural modeling projects that wish to open themselves up for collaboration with the rest of the academic community. OSB technology is built around the NeuroML v2 data format for neural model specification. OSB is not structured around single simulation runs but around projects (which loosely correspond to single models), for which it provides a front-end web page listing some essential information (e.g., project description, members, references, etc.) and a link to the project's code repository (e.g., GitHub). For a project that is not converted to the NeuroML format, this is all information that is directly introspectable from OSB. For such projects, OSB essentially provides a centralized space where model authors can build a web page about their project. Additionally, for parts of the project that are converted to NeuroML, one can invoke the Geppetto (http://www.geppetto.org/) Java interface within the web browser, allowing the user to inspect the model in detail via a GUI. One limitation of this approach is that NeuroML has been designed for detailed morphological neural models, and large-scale point neuron simulations, common in systems neuroscience, are not as well supported. Unlike Arkheia, OSB does not offer an explicit formalized presentation of the simulation, results, experimental protocols, and their parametric context, which we argue are key for further development of collaborative tools in computational neuroscience.
Architecture
Arkheia follows the standard database-server-client architecture. To facilitate both efficient and flexible storage of complex highly structured data that describe the makeup and results of neural simulations, we have selected the modern document-based database MongoDB (https://www.mongodb.com/). The nature of the data describing a simulation run and its results are straightforwardly described by a hierarchical document that can be efficiently represented and retrieved in a document-based database. Presently, MongoDB represents an industry standard for document-based databases and is particularly used frequently in web-based software solutions. It is well-supported and accessible to new users, overall making it a suitable choice for this project.
The data stored in the database is served to the client via a thin server layer developed in the the asynchronous event-driven JavaScript runtime Node.js (https://nodejs.org/) using the Express.js (https://expressjs.com/) web-server package. The client is a web application written in the AngularJS (https://angularjs.org/) framework (see Figure 1). The server uses the Mongoose library to access and standardize the data stored in the MongoDB instance. The architecture of the client follows the Model-View-Controller (MVC) design facilitated by the AngularJS framework. Here the different Angular models map onto different parts of the simulation run description (i.e., the root list of simulation runs, stimuli, results, experiment protocols, etc.), and each Angular model is associated with an HTML template and a controller handling the dynamic aspects of the views. The client is a multi-page web application offering multiple views of the simulation run data, mostly following a tabular presentation pattern. Additionally, a more complex web application for interactive exploration of parameter search results is offered.
|
The insertion of data into Arkheia is expected to be done by an arbitrary set of back-ends, which should automatically export data from a given simulation framework and insert it into the MongoDB database in the format expected by Arkheia, which is described in the following section. The back-ends connect directly to the database, and thus no assumptions about their behavior beyond the insertion of the data in the correct format are made. The interaction of Arkheia with external tools is thus fully specified by the format of data stored in the database. While the back-ends are primarily meant as automatic exporters from simulation environments, in principle one could create an interactive GUI-based application (which from the point of view of Arkheia would behave as any other back-end) that would allow manual insertion of data into Arkheia.
API
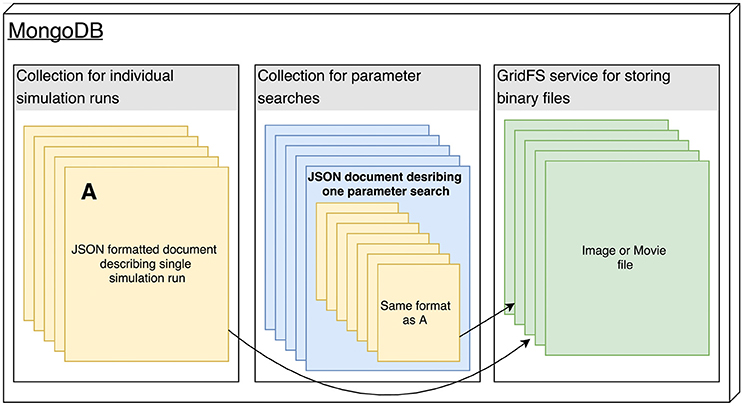
The Arkheia API is essentially a description of how the shared data about individual simulation runs should be stored in the MongoDB database used by Arkheia (see Figure 2). The data is stored in three MongoDB collections, one storing the individual simulation runs, one storing the parameter searches and one storing any binary files (e.g., images and movies) that are referenced from the documents in the other two collections (see Figure 2). Thus, with the exception of the mechanisms for storing of image and movie data, this storage description reduces to the description of the format of the hierarchical document that will be stored for each simulation run. This specification covers the storage of model specification, sensory stimuli, experimental protocols, resulting data analysis, and visualization outputs. We expect rapid development in the specifications of data Arkheia handles, both as the scope of Arkheia expands, but more importantly as we hope standardized specifications of some aspects of the data will develop in the field in the near future.[24]
|
Simulation run representation
Arkheia's data specification follows a document-based design, which conveniently maps onto the document-based MongoDB database used by Arkheia. Each simulation run is represented by a single JSON hierarchical data structure which corresponds to the document inserted into the database. Thus the Arkheia input data specification reduces to the expected format of this JSON data-structure. At the root level, the SimulationRun data-structure contains the following entries with the indicated value types:
{
’submission_date’ : string,
’run_date’ : string,
’simulation_run_name’ : string,
’model_name’ : string,
’results’ : list of Result,
’stimuli’ : list of Stimulus,
’recorders’ : list of Recorder,
’experimental_protocols’ : list of Protocol,
’parameters’ : ParameterSet
}
The submission_date and run_date entries are expected to be strings representing time in “YYYY/MM/DDHH:MM:SS” format. The simulation_run_name is an arbitrary name given to this specific simulation run (not the model). The model_name is the name of the model that was simulated. The results, stimuli, recorders, and experimental_protocols are each a list of JSON data structures, the format of which will be described below. Finally the parameters variable should describe the full parametrization of the model used in this run, and as all parametrization throughout Arkheia API, it should follow the ParameterSet format.
ParameterSet is a nested dictionary (see schema below) where each value associated with a key (that corresponds to the name of the parameter) is a tuple (a, b, c), where a corresponds to the value of the parameter and can either be a scalar or ParameterSet itself, b is the type of the parameter, and c is a short description of the parameter's meaning.
ParameterSet = {
’key’ : (scalar value, scalar type,
description string),
or
’key’ : (ParameterSet, dict,
. description string)
.
.
.
}
The results root-level entry should contain a list of Result JSON data-structures, each describing one result produced during the simulation (presumably after analysis and visualization of the raw data recorded during the simulation). Each result is meant to be represented as a figure with an accompanying explanatory caption and is represented as the following JSON data-structure:
{
’code’ : string,
’name’ : string,
’caption’ : string,
’parameters’ : ParameterSet,
’figure’ : MongoDB GridFS ID
}
The code entry should contain a reference, for example a fully qualified class or function name, to the source code that generates the given figure. The name should contain the name of the figure, and the caption should contain a caption that describes what is displayed in the figure. The parameters entry should contain the parameters with which the generator of the figure identified by code entry was invoked to generate the given figure, and it should follow the ParameterSet format described above. Finally, the figure entry contains a reference to the image of the figure (in any format that is widely supported in a browser) that was stored in the MongoDB GridFS file store.
The stimuli root-level entry should contain a list of Stimulus JSON data-structures, each describing one of the stimuli presented during the simulation. It assumes that the model was presented with a list of stimuli, generated by some source code entity (class or function) and parameters, and its “raw” instantiation can be represented as a vector stream, which in turn can be translated into a movie for visual inspection by the user. The format of each Stimulus data-structure is as follows:
{
’code’ : string,
’short_description’ : string,
’long_description’ : string,
’parameters’ : ParameterSet,
’movie’ : MongoDB GridFS ID,
}
The code and parameters entries are analogous to those in the Result data structure. Entries short_description and long_description, respectively, should contain a brief (one sentence) and a more detailed description of the stimulus. The movie entry should contain a reference to the movie (animated gif) of the stimulus (stored in the MongoDB GridFS).
The recorders root-level entry should contain information on the recording configuration present during the simulation. It assumes that this can be described as a list of parametrized entities that each records some set of variables from some set of neurons. The recorders entry should thus contain a list of Recorder JSON data structures, each corresponding to one such recording configuration entity. The format of the Recorder data structure is as follows:
{
’code’ : string,
’short_description’ : string,
’long_description’ : string,
’parameters’ : ParameterSet,
’variables’ : list of strings,
’source’ : string
}
The code, parameters, short_description, and long_description entries are analogous to those in the Stimulus data structure. The variables entry should contain a list of strings, each identifying the variable that the given recording configuration recorded in the selected neurons, and the source should contain the name of the population of neurons to which this recording configuration was applied.
Finally the experimental_protocols root-level entry is expected to contain a list of Protocol JSON data structures characterizing the experimental protocols that were performed during the simulation of the model. The format of each of the Protocol data structures is as follows, and the semantics of all the parameters are analogous to the corresponding entries in the Stimulus data structure:
{
’class’ : string,
’short_description’ : string,
’long_description’ : string,
’parameters’ : ParameterSet
}
Parameter search representation
Parameter search is essentially a collection of simulation runs with a systematically varying subset of parameters. In principle, it would be possible to add the simulation runs originating from parameter searches into the same database as the individual simulation runs, and keep a separate record of their membership to the parameter search group. We have decided against such an organization, as parameter searches can consist of hundreds or more runs and would thus clog the presentation of the individual runs, as well as potentially make access to the individual run database slow. The parameter searches are thus stored in a separate database (within the same instance of the MongoDB server). Each parameter search is a JSON document of the following format:
{
’submission_date’ : string,
’name’ : string,
’simulation_runs’ : list of SimulationRun,
’parameter_combinations’ : list of tuples
}
Whereas submission_date is expected to be a string containing the data of the submission to the repository, the name entry is the name given to the simulation run. The simulation_runs entry should contain a list of SimulationRun JSON data-structures, each corresponding to one simulation run with the same format as the JSON data-structure describing an individual simulation run described in the previous section. Finally the parameter_combinations entry should contain a list of tuples, with each tuple holding the name of the parameter that was varied, and a list of parameter values that were explored (note that currently Arkheia only supports grid parameter searches, although irregular grids and missing elements are both supported).
API design discussion
We want to first emphasize that the goal of Arkheia is not to provide storage for the full raw data associated with a given simulation. Rather, it provides a structured description of the simulation context and results, sufficient for replication or reuse of the various aspect of the given simulation, and it facilitates a convenient human readable comparison and browsing of models and their results.
The data format specification described above broadly follows the organization of data in the Mozaik framework—facilitating seamless integration with the workflow toolkit—but is designed with sufficient flexibility to allow integration with other modeling frameworks. The specification makes a number of assumptions about the nature of data, describing the simulation run and its results, which, however, we believe are mappable onto the majority of use cases in systems neuroscience.
For example, we assume that the full parameterization of the model can be expressed as a tree structure, or more precisely a forest, as we allow multiple root entries. Note that the most common format for simulation configuration, a plain list of parameter names and their values, is trivially mapped onto the forest structure as a forest of depth one, while we are not aware of any major simulation configuration schemes that cannot be mapped straightforwardly onto such a tree structure. If the configuration involves binary data (e.g., a specific image used for stimulation, or some data derived from biological experiments that set specific neuron-to-neuron connectivity), these can be simply referenced by the name of the file containing the binary data, which can be later—if necessary—looked up in an external repository holding the full raw data of the given simulation run.
Another general approach within the specification securing flexibility and generality is the assumption that the various entities to be shared are generated by well-defined blocks of code that can be easily referenced (i.e., classes or functions) and thus by sharing the reference to this code and the parametrization used in the given specific simulation run, one can fully recreate the entity, providing one has access to the full source code of the given simulation framework (and any potential additions to it by the author of the simulation). Note that the latter is a reasonable assumption under the condition that the author is willing to publish the model in Arkheia and should in any case become a standard practice in the age of open science. These code entities should furthermore be accompanied by a detailed description of the given entity (e.g., experimental protocol), with an explanation of how the associated parameters configure the entity. An elegant solution to this is to keep this description as a code “docstring” of the given code block, where it fulfills the good practice of well-documented code, and can be automatically harvested into the documentation of the given simulation framework while at the same time exported for publication in Arkheia.
The final source of flexibility is that most of the entries of the specification do not actually have to hold data (e.g., all the string values can be set to empty strings, or list values to empty lists). Arkheia can thus be used in projects which either do not use or do not yet expose all the data so far covered in our specification. It is the intention that this specification will be developed further, with the main goal of increasing the coverage of the information that can be exposed about simulation runs. We particularly hope that some of the ongoing efforts in the wider community will generate well-designed and popular specification standards for some of the aspects of neural simulations covered by Arkheia (such as standardization of experimental protocols, higher level model specifications, or stimulus definitions), which we would eagerly seek to incorporate into Arkheia.[25] To this end, Arkheia represents both a sketch of how such specifications could look and an example of how they could be used to facilitate communication and comparison across different models, thus motivating the development of these technologies, which we believe are key to the future computational neuroscience software infrastructure.
Back-end implementation
The API presented above fully specifies the requirements Arkheia imposes upon its back-ends, and thus all remaining implementation choices are left to the author of the given back-end. However, to provide reader a basic understanding of what it takes to implement such a back-end, we will in this section present a brief account of the Mozaik back-end that currently ships with Arkheia.
As a part of a simulation run, Mozaik creates a new directory, in Mozaik referred to as data-store, in which it stores the recorded neural signals coupled to a detailed specification of the simulation context. The role of the Mozaik Arkheia back-end is to simply transform this data-store (ignoring some aspects of it not covered in Arkheia such as raw neural signal recordings) into the Arkheia document format specified in this section above and submit it into a user-specified Arkheia repository.
The Mozaik Arkheia back-end is written in Python and uses the pymongo package to access and transmit the Arkheia document into the requested Arkheia repository, taking care of all the low-level MongoDB related processing. Thus, the MongoDB-specific code within the back-end amounts to only six lines of code (at the time of writing). We expect a similar level of complexity for other programming languages subject to the availability of a similarly functional MongoDB library.
The remaining work of the back-end is relegated to creating a nested Python dictionary, restricted to elementary scalar types (i.e., float, int, string) or arrays of them as leafs, that follows the Arkheia document specification and reflects the information contained in the Mozaik data-store. Given the explicit exposure of all the required information in Mozaik, this amounts to simple systematic browsing through the data-store, using the access routines provided by Mozaik, and, subject to few simple formatting translations, re-inserting the information into the API pre-specified format. The one extra processing step the Mozaik back-end undertakes is that using the code-references (in the form of full path to generating class) of the various elements of the simulation context, it automatically harvests the values for the short and long description fields (see the API description above) from the “docstrings” of the referenced code. This is possible because of the docstring formatting convention (required by the Sphynx [1] documentation package) that Mozaik adheres to. This Mozaik-specific design choice saves time by reusing the information entered by the user during documentation of the code also for the purpose of serving via Arkheia. All-in-all, the entire back-end is thus only a very modest 300 lines of code, including all the auxiliary routines.
In terms of usage, the Mozaik back-end is invoked via command-line, with two parameters: the path to the data-store previously created by Mozaik simulation run, and the address of the Akrheia repository to which the user wishes to submit the results. It should be noted that we have chosen this command-line-centric usage for its flexibility, but in principle the invocation of the back-end could for example be directly integrated into the Mozaik platform so that it automatically happens at the end of each simulation run, or integrated into any GUI, if available for the simulation platform. Ultimately this design choice is left to be made by authors of any specific back-end.
The amount of work required to implement a back-end will naturally vary between simulation platforms, depending mainly on how explicitly represented and easy-to-retrieve the various information that Arkheia serves are in the given simulation platform. However, here we demonstrate, that for those platforms built with the explicit coupling of in-silico recordings with the information about simulation context in mind, this can be a very straightforward process.
Web-based graphical front-end
References
- ↑ Markram, H. (2006). "The blue brain project". Nature Reviews Neuroscience 7 (2): 153–60. doi:10.1038/nrn1848. PMID 16429124.
- ↑ Rangan, A.V.; Tao, L.; Kovacic, G.; Cai, D. (2009). "Multiscale modeling of the primary visual cortex". IEEE Engineering in Medicine and Biology Magazine 28 (3): 19–24. doi:10.1109/MEMB.2009.932803. PMID 19457730.
- ↑ Markram, H.; Meir, K.; Lippert, T. et al. (2011). "Introducing the Human Brain Project". Procedia Computer Science 7: 39–42. doi:10.1016/j.procs.2011.12.015.
- ↑ Koch, C.; Reid, R.C. (2011). "Neuroscience: Observatories of the mind". Nature 483 (7390): 397–8. doi:10.1038/483397a. PMID 22437592.
- ↑ Bouchard, K.E.; Aimone, J.B.; Chun, M. et al. (2016). "High-Performance Computing in Neuroscience for Data-Driven Discovery, Integration, and Dissemination". Neuron 92 (3): 628-631. doi:10.1016/j.neuron.2016.10.035. PMID 27810006.
- ↑ Hawrylycz, M.; Anastassiou, C.; Arkhipov, A. et al. (2016). "Inferring cortical function in the mouse visual system through large-scale systems neuroscience". Proceedings of the National Academy of Sciences of the Unites States of America 113 (27): 7337–44. doi:10.1073/pnas.1512901113. PMC PMC4941493. PMID 27382147. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4941493.
- ↑ Anderson, D.P.; Cobb, J.; Korpela, E. et al. (2002). "SETI@home: An experiment in public-resource computing". Communications of the ACM 45 (11): 56–61. doi:10.1145/581571.581573.
- ↑ Szigeti, B.; Gleeson, P.; Vella, M. et al. (2014). "OpenWorm: An open-science approach to modeling Caenorhabditis elegans". Frontiers in Computational Neueroscience 8: 137. doi:10.3389/fncom.2014.00137. PMC PMC4217485. PMID 25404913. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4217485.
- ↑ Carnevale, N.T.; Hines, M.L. (2006). The Neuron Book. Cambridge University Press. pp. 480. ISBN 9780521843218.
- ↑ Gewaltig, M.-O.; Diesmann, M.. "NEST (NEural Simulation Tool)". Scholarpedia 2 (4): 1430. doi:10.4249/scholarpedia.1430.
- ↑ Stimberg, M.; Goodman, D.F.; Benichoux, V.; Brette, R.. "Equation-oriented specification of neural models for simulations". Frontiers in Neuroinformatics 8: 6. doi:10.3389/fninf.2014.00006. PMC PMC3912318. PMID 24550820. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3912318.
- ↑ Bekolay, T.; Bergstra, J.; Hunsberger, E. et al.. "Nengo: a Python tool for building large-scale functional brain models". Frontiers in Neuroinformatics 7: 48. doi:10.3389/fninf.2013.00048. PMC PMC3880998. PMID 24431999. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3880998.
- ↑ Davison, A.P.; Brüderle, D.; Eppler, J. et al.. "PyNN: A Common Interface for Neuronal Network Simulators". Frontiers in Neuroinformatics 2: 11. doi:10.3389/neuro.11.011.2008. PMC PMC2634533. PMID 19194529. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2634533.
- ↑ Garcia, S.; Guarino, D.; Jaillet, F. et al.. "Neo: An object model for handling electrophysiology data in multiple formats". Frontiers in Neuroinformatics 8: 10. doi:10.3389/fninf.2014.00010. PMC PMC3930095. PMID 24600386. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3930095.
- ↑ 15.0 15.1 Stevens, J.L.; Elver, M.; Bednar, J.A.. "An automated and reproducible workflow for running and analyzing neural simulations using Lancet and IPython Notebook". Frontiers in Neuroinformatics 7: 44. doi:10.3389/fninf.2013.00044. PMC PMC3874632. PMID 24416014. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3874632.
- ↑ 16.0 16.1 Meyer, R.; Obermayer, K.. "pypet: A Python Toolkit for Data Management of Parameter Explorations". Frontiers in Neuroinformatics 10: 38. doi:10.3389/fninf.2016.00038. PMC PMC4996826. PMID 27610080. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4996826.
- ↑ Friedrich, P.; Vella, M.; Gulyás, A.I. et al.. "A flexible, interactive software tool for fitting the parameters of neuronal models". Frontiers in Neuroinformatics 8: 63. doi:10.3389/fninf.2014.00063. PMC PMC4091312. PMID 25071540. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4091312.
- ↑ Sobolev, A.; Stoewer, A.; Pereira, M. et al.. "Data management routines for reproducible research using the G-Node Python Client library". Frontiers in Neuroinformatics 8: 15. doi:10.3389/fninf.2014.00015. PMC PMC3942789. PMID 24634654. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3942789.
- ↑ Pröpper, R.; Obermayer, K.. "Spyke Viewer: A flexible and extensible platform for electrophysiological data analysis". Frontiers in Neuroinformatics 7: 26. doi:10.3389/fninf.2013.00026. PMC PMC3822898. PMID 24273510. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3822898.
- ↑ Thibeault, C.M.; O'Brien, M.J.; Srinivasa, N.. "Analyzing large-scale spiking neural data with HRLAnalysis". Frontiers in Neuroinformatics 8: 17. doi:10.3389/fninf.2014.00017. PMC PMC3942659. PMID 24634655. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3942659.
- ↑ Woodman, M.M.; Pezard, L.; Domide, L. et al.. "Integrating neuroinformatics tools in TheVirtualBrain". Frontiers in Neuroinformatics 8: 36. doi:10.3389/fninf.2014.00036. PMC PMC4001068. PMID 24795617. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4001068.
- ↑ Kubilius, J.. "A framework for streamlining research workflow in neuroscience and psychology". Frontiers in Neuroinformatics 7: 52. doi:10.3389/fninf.2013.00052. PMC PMC3894454. PMID 24478691. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3894454.
- ↑ 23.0 23.1 Antolík, J.; Davison, A.P.. "Integrated workflows for spiking neuronal network simulations". Frontiers in Neuroinformatics 7: 34. doi:10.3389/fninf.2013.00034. PMC PMC3857637. PMID 24368902. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3857637.
- ↑ Eglen, S.J.; Marwick, B.; Halchenko, Y.O. et al.. "Toward standard practices for sharing computer code and programs in neuroscience". Nature Neuroscience 20 (6): 770–3. doi:10.1038/nn.4550. PMID 28542156.
- ↑ Hucka, M.; Finney, A.; Sauro, H.M. et al.. "The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models". Bioinformatics 19 (4): 524–31. PMID 12611808.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. The original article lists references alphabetically, but this version — by design — lists them in order of appearance. What were originally footnotes have been turned into inline external links.