Journal:Assessment of and response to data needs of clinical and translational science researchers and beyond
| Full article title | Assessment of and response to data needs of clinical and translational science researchers and beyond |
|---|---|
| Journal | Journal of eScience Librarianship |
| Author(s) | Norton, Hannah F.; Tennant, Michele R.; Botero, Cecilia; Garcia-Milian, Rolando |
| Author affiliation(s) | University of Florida, Yale University |
| Primary contact | Email: nortonh at ufl dot edu |
| Year published | 2016 |
| Volume and issue | 5 (1) |
| Page(s) | e1090 |
| DOI | 10.7191/jeslib.2016.1090 |
| ISSN | 2161-3974 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://escholarship.umassmed.edu/jeslib/vol5/iss1/2/ |
| Download | http://escholarship.umassmed.edu/cgi/viewcontent.cgi?article=1090&context=jeslib (PDF) |
Abstract
Objective and setting: As universities and libraries grapple with data management and “big data,” the need for data management solutions across disciplines is particularly relevant in clinical and translational science research, which is designed to traverse disciplinary and institutional boundaries. At the University of Florida Health Science Center Library, a team of librarians undertook an assessment of the research data management needs of clinical and translation science (CTS) researchers, including an online assessment and follow-up one-on-one interviews.
Design and Methods: The 20-question online assessment was distributed to all investigators affiliated with UF’s Clinical and Translational Science Institute (CTSI) and 59 investigators responded. Follow-up in-depth interviews were conducted with nine faculty and staff members.
Results: Results indicate that UF’s CTS researchers have diverse data management needs that are often specific to their discipline or current research project and span the data lifecycle. A common theme in responses was the need for consistent data management training, particularly for graduate students; this led to localized training within the Health Science Center and CTSI, as well as campus-wide training. Another campus-wide outcome was the creation of an action-oriented Data Management/Curation Task Force, led by the libraries and with participation from Research Computing and the Office of Research.
Conclusions: Initiating conversations with affected stakeholders and campus leadership about best practices in data management and implications for institutional policy shows the library’s proactive leadership and furthers our goal to provide concrete guidance to our users in this area.
Keywords: needs assessment, clinical and translational science, service development
Objective and settings
Biomedical researchers work with considerable amounts of heterogeneous data; managing these datasets raises new challenges in terms of acquiring, archiving, annotating, and analyzing data. Libraries across the nation and the world are developing tools to manage this research data, extending natural skills within libraries for organizing, sharing, and archiving information, as well as educating staff about best practices. This stems largely from an increased interest in data management and data sharing at the researcher level, fueled by both funders’ inclusion of data management plan requirements in proposals and by collaborative, large-scale research projects that generate data that is “big” and diverse.[1] The need for data management solutions across disciplines is particularly relevant in clinical and translational science (CTS) research, which is designed to cut across disciplinary and institutional boundaries. Data sharing, organization, storage, and security must scale up to meet these growing needs.
A number of roles in data management and curation have been proposed for librarians including, among others: hosting institutional and disciplinary repositories, developing data publication standards, supporting documentation and metadata use, training researchers and students in funders’ requirements and best practices in data management, working more directly with offices of research, deploying existing tools, hosting data management events (symposia, reflective workshops), embedding into research laboratories to provide data management solutions, and advocating for data sharing.[2][3][4][5][6][7][8][9][10][11][12] Reznick-Zellen et al.[13] postulate three “tiers” of library-based data management services: education (for example, LibGuides, webpages, and workshops), consultation (on data management plans, metadata standards, repository deposition, etc.), and infrastructure (data staging platforms and repositories).
With limited resources available, an integral step to developing these new services is identifying specific needs of the patrons to whom these services are targeted and ensuring that time and resources go into services that truly map to those needs. Needs assessments can also illuminate issues outside of the scope of direct library services, but for which librarians can be advocates on the institutional level. Although the importance of needs assessment is widely agreed upon[14] and a number of libraries have performed such assessments of data management needs[15][16][17][18][19][8][11][20], a 2009 survey of ARL institutions indicated that 62% of responding institutions had not performed a data needs assessment although 73% of libraries had some involvement in e-Science at their institution.[21]
Beginning in 2006, the National Institutes of Health (NIH) began offering Clinical and Translational Science Awards (CTSAs) to institutions across the country in order to minimize the time from discovery to clinical practice, enhance community-engagement in clinical research, and train new clinical and translational science researchers.[22] In 2009, the University of Florida (UF) received CTSA funding for its existing Clinical and Translational Science Institute (CTSI). As of 2015, the CTSI’s reach has expanded to more than 1,800 investigators across the University’s 16 colleges using CTSI services.[23]
The UF Health Science Center Library (HSCL) serves the six colleges of UF’s Academic Health Center (Dentistry, Medicine, Nursing, Pharmacy, Public Health and Health Professions, and Veterinary Medicine) and related centers and institutes, including the CTSI. HSCL is part of the broader campus library system, the George A. Smathers Libraries. At HSCL, dual interests in campus researchers’ data management needs and those particular to the CTSI led a team of librarians to undertake an assessment of the research data management needs of CTS researchers, including an online assessment and follow-up, one-on-one interviews. This assessment was situated within a broader project funded by the National Network of Libraries of Medicine, Southeast Atlantic Region focused on assessing CTS researcher needs: general information needs, bioinformatics needs, and data needs. Given the diversity of CTS researchers and the centrality of data to their research, HSCL librarians identified CTSI-affiliated researchers as an ideal pilot group to use for campus data needs assessments. At the same time, HSCL librarians developed a strong partnership with the Director of UF’s High Performance Computing Center (now known as Research Computing), who values the library’s role in data endeavors. He joined two of the Smathers Libraries’ Associate Deans (including author CB) in participating in the ARL E-Science Institute in 2011 and performing a campus environmental scan related to e-science and data services focused primarily on the plans and attitudes of high-level administrators. Additional suggestions for service development were gathered when three of the authors (CB, MRT, HFN) used funding awarded through UF’s Faculty Enhancement Opportunity program (mini-sabbaticals) to visit Purdue University’s library and learn from its successful data program.
Design and methods
The authors conducted a multimodal needs assessment using a combination of an online survey and in-depth, one-on-one semi-structured interviews. Semi-structured interviews were selected as a complementary means of data collection because they are well suited for exploring respondents’ perceptions and opinions on complex issues. In addition, they enable asking for more information and clarification of answers.[24] In order to ensure the safety of study participants and confidentiality of their data, both the survey and the subsequent interviews were approved by the University of Florida Institutional Review Board (Exemption #U-1142-2011).
Survey
In the spring of 2012, a team of three HSCL librarians distributed a 20-question online assessment (see Appendix 1) to all investigators affiliated with UF’s Clinical and Translational Science Institute, a total of 834 individuals. Questions were developed in collaboration with the director of UF’s High Performance Computing Center and colleagues in the main campus library’s Digital Library Center.
Interviews
In order to obtain more in-depth information from a subset of individuals across the CTSI, three HSCL librarians conducted interviews with CTSI-affiliated faculty or staff. The full list of CTSI-affiliated researchers was reviewed by librarian team members, and 58 individuals were identified who had worked closely with the libraries in the past and represented diverse disciplines; these individuals were contacted about participating in interviews. Nine individuals from this list agreed to be interviewed. Each interview lasted 30-60 minutes and was audio-recorded for later transcription and qualitative coding into themes; all interviews were conducted by two librarians (with one exception in which only one librarian conducted the interview). The interviews were organized around a series of questions modified from the University of Virginia Libraries’ data interview template, which itself is modified from Purdue’s Data Curation Profile interview template.[16] These questions addressed the broad topics of research area, data types, how data is worked with, preservation concerns, sharing and long-term accessibility, and what assistance from the library or other campus entities would make data management easier (see Appendix 2). The interview format was flexible enough that participants were able to address any arising concerns or comments about data management that did not fit into these categories. The invitation to participate in interviews and the in-person introduction on the day of the interview stressed that the interview was part of a broad needs assessment regarding data management and that any related concerns or barriers could be discussed. All of the authors sequentially reviewed the interview transcripts, identified relevant quotes, and coded them using 21 themes (e.g. sharing, backups, lab notebooks, etc.).
Results
Survey
Fifty-nine investigators responded to the survey, for a response rate of 7.1 percent. Survey respondents represented nine of UF’s 16 colleges, with a majority of responses coming from five of the six Health Science Center colleges served directly by the HSCL: Medicine (59.3 percent), Public Health & Health Professions (9.3 percent), Dentistry (7.4 percent), Pharmacy (5.6 percent), and Veterinary Medicine (1.9 percent). Other colleges represented were Agriculture and Life Sciences (7.4 percent), Liberal Arts & Sciences (3.7 percent), and Journalism (1.9 percent). The vast majority of respondents were faculty members (93.2 percent); the remainder were graduate students (3.4 percent), postdocs (1.7 percent), and staff (1.7 percent).
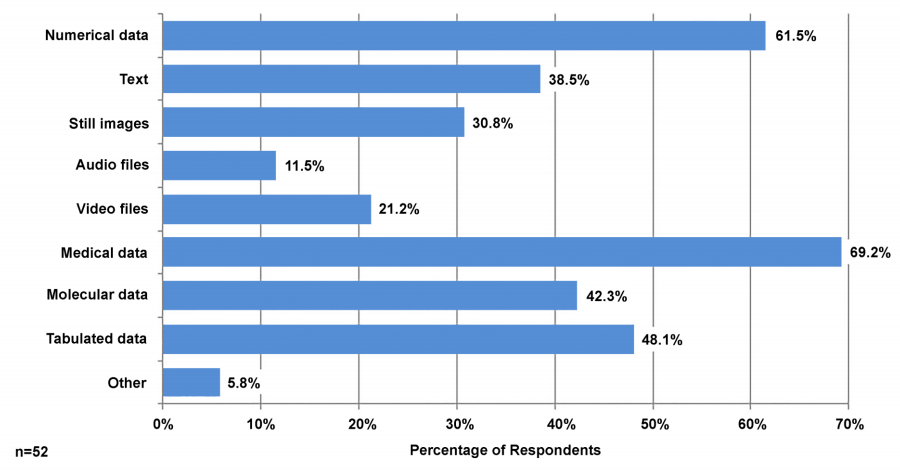
Figure 1 shows the types of data that survey respondents said they generate. Respondents could choose as many data types as were relevant to them, and on average they listed at least three types of data. The most commonly chosen types of data were medical (69.2 percent), numerical (61.5 percent), tabulated (48.1 percent), molecular (42.3 percent), and text data (38.5 percent). Mentioned under “other data” were qualitative data, performance data, and MRI images.
Participants were asked to list the formats in which their data exist (what file formats or file extensions they use); this open-text question had a lower response than the multiple-choice questions (n=29). The overwhelming majority of respondents use spreadsheets (82.8 percent). Other frequently mentioned file formats were those for specific statistical software (34.5 percent), word processing documents (27.6 percent), images (24.1 percent), databases (20.7 percent), and other file formats (24.1 percent) followed by video (13.8 percent) and text (6.9 percent). Other formats listed included audio, code, survey responses, and PowerPoint. This frequent use of non-specific applications such as spreadsheets and word processing documents mirrors results elsewhere in the literature.[15]
|
Participants were asked how their data are labeled or annotated and then asked to select as many of the four options as applied to them. Many respondents were performing (or having someone on their research team create) manual annotation (78.8 percent); 32.7 percent were generating labels automatically through a data collection tool; 21.2 percent were using a codebook to annotate referentially; and 17.3 percent of respondents indicated that their data are not annotated.
|
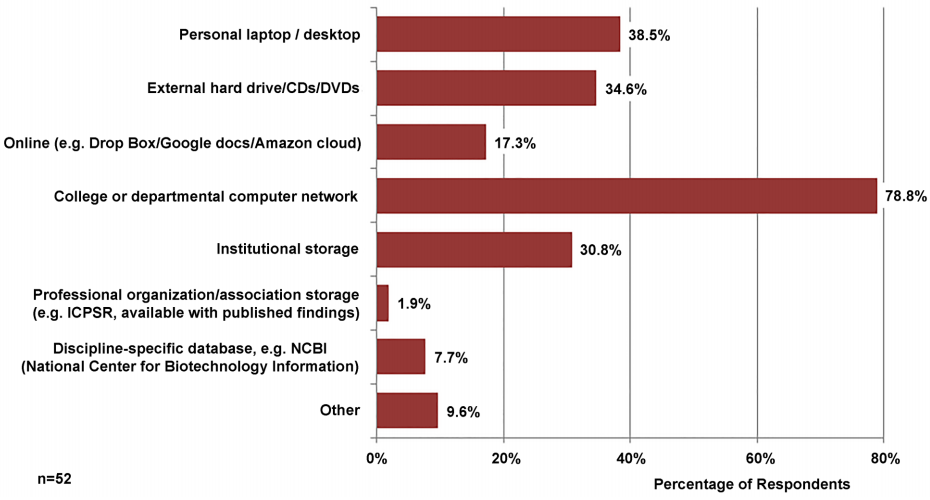
Participants were asked how they store their data; their responses are reported in Figure 2. Respondents could choose multiple methods, and on average respondents used at least two of the methods listed. Highly localized options included personal laptop or desktop (38.5 percent) and external hard drive or CDs or DVDs (34.6 percent). Institution-specific storage options were the most popular with 78.8 percent of respondents using a college or departmental computer network and 30.8% using institutional storage. Least popular were national-level, discipline repositories including professional organization or association storage (1.9 percent) and discipline-specific databases (7.7 percent). Although data later in this survey indicates that more participants were using discipline-specific repositories for sharing, this data suggests that participants did not consider these repositories a storage solution. Other types of storage mentioned were secure online databases including REDCap.[25]
|
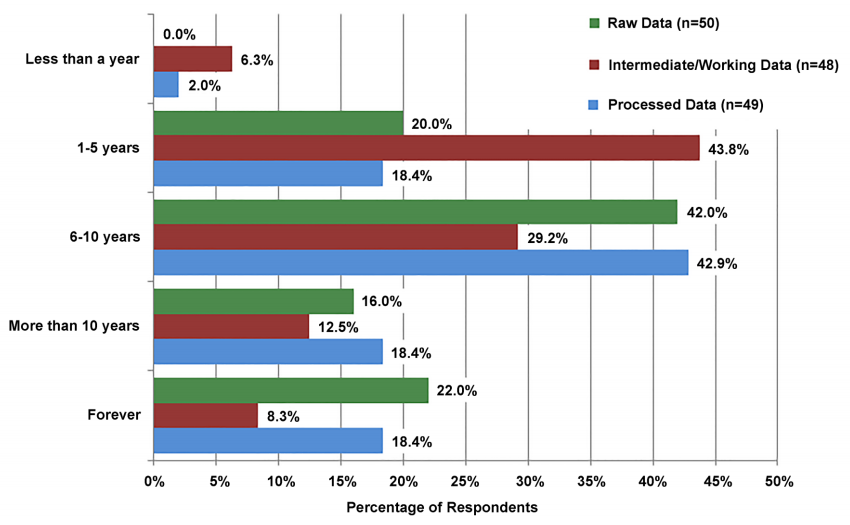
Participants were asked how long they need their data stored, with raw, intermediate, or working data, and processed data considered separately. Figure 3 shows these results. Most responses fell into the categories of 1-5 years and 6-10 years. Very few respondents indicated that any data should be kept less than a year (none for raw data, 6.3 percent for intermediate/ working data, 2.0 percent for processed data). The most commonly desired storage time for intermediate/working data was 1-5 years (43.8 percent); the number of respondents choosing subsequent time periods decreased for each longer time period (29.2 percent wanting to keep it for 6-10 years, 12.5 percent for more than 10 years, and 8.3 percent forever). In contrast, the most commonly desired storage time for processed data was 6-10 years (42.9 percent), with an even split (18.4 percent) of respondents wanting to keep it for 1-5 years, for more than 10 years, or forever. Raw data was most commonly kept for 6-10 years (42.0 percent), with 20.0 percent of respondents wanting to keep it for 1-5 years, 16.0 percent for more than 10 years, and 22.0 percent forever.
Participants were asked who they are willing to share data with; responses roughly indicate that the closer to their work, the more likely researchers are willing to share. The survey showed 95.8 percent of respondents are willing to share with their immediate collaborators; 35.4 percent with others in their department or institute; 35.4 percent with others in their disciplines; 16.7 percent with others outside of their field; and 6.3 percent with anyone.
|
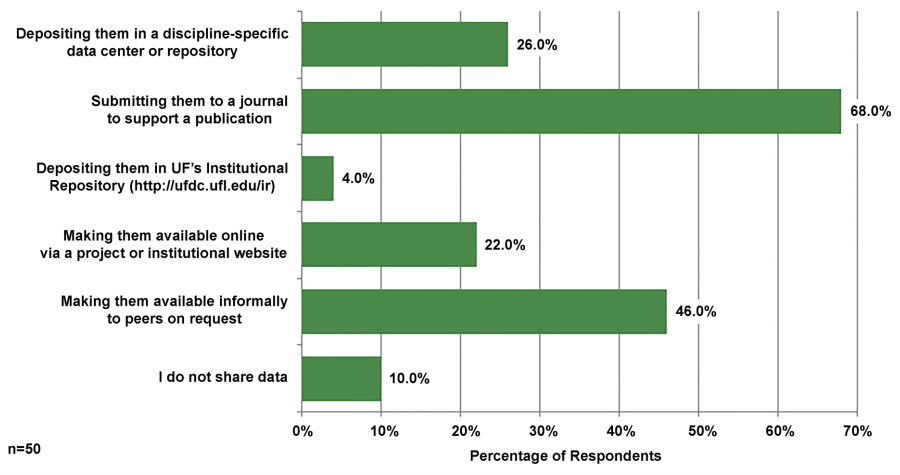
Participants were asked how they were sharing or planning to share their data (see Figure 4). The most common responses were submitting them to a journal to support a publication (68.0 percent) and making them available informally to peers on request (46.0 percent). Some respondents indicated that they shared by depositing data in a discipline-specific data center or repository (26.0 percent) or making them available online via a project or institutional website (22.0 percent). Only 4.0 percent of respondents indicated that they shared data by depositing them to UF’s Institutional Repository; 10.0 percent of respondents indicated that they do not share data.
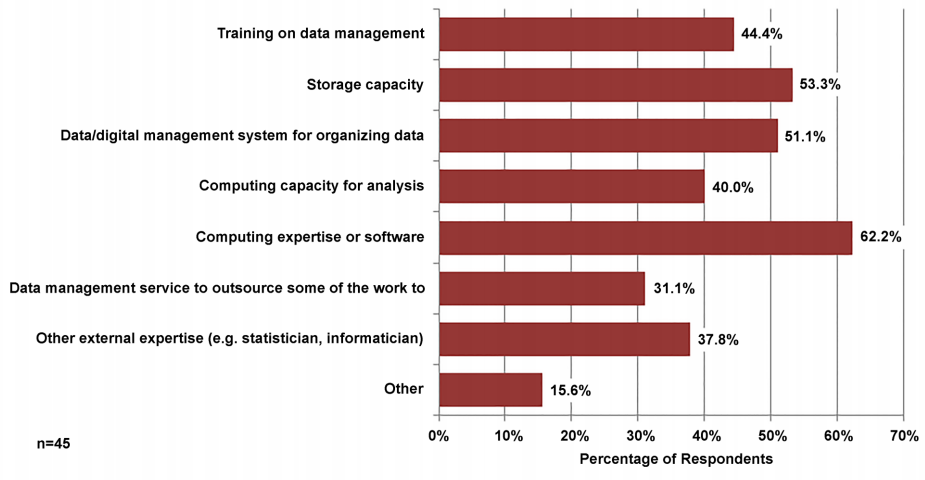
Participants were asked what resources outside their department they needed to best manage and analyze their data (see Figure 5). The most frequently mentioned responses deal with technical needs for computing expertise or software (62.2 percent) and storage capacity (53.3 percent). Other popular responses were a data/digital management system for organizing data (51.1 percent), training on data management (44.4 percent), and computing capacity for analysis (40.0 percent). Some respondents also identified other external expertise such as a statistician or an informatician (37.8 percent) or a data management service to outsource some of the work to (31.1 percent) as needed. Other needs mentioned included network security and statistical software.
|
Interviews
The nine data interviews were conducted with participants from five of UF’s 16 colleges (Agriculture & Life Sciences, Medicine, Pharmacy, Public Health & Health Professions, Veterinary Medicine). Eight of the interviews were with faculty members, and one was with a staff member; in one case a graduate student participated in the interview with his faculty advisor. Table 1 provides a summary of the affiliation of interviewees, types of research they perform, and types of data they generate. Several of the most commonly addressed themes from the interviews are addressed below.
| ||||||||||||||||||||||||||||||||||||||||||||
Across interviews, participants noted a lack of consistency in data management practices, based in large part on minimal or ad hoc training available to both students and faculty on data management. Interviews revealed that graduate students currently learn data organization and management informally, either from PIs or on their own; this finding corroborates the findings of Peters and Vaughn[8] that graduate students are rarely formally taught data management competencies. As one participant noted, “I think right now it’s kind of a crash course for graduate students…Because no one teaches you how to organize data. It starts to accumulate and accumulate and accumulate, and you just have all these files and you say, I don’t know.” This can cause problems for individuals and is a perennial problem for larger labs with many graduate students each storing, organizing, and documenting data in their own way, especially when a student or postdoc leaves the lab and others need to use his or her data, as noted elsewhere in the literature.[11] Participants largely agreed that more systematic training would be helpful. When asked about unmet needs, one faculty member suggested, “… more training of graduate students for how to put together data sets and what to be aware of, and what resources are available.” Faculty also generally learn data management through trial and error and self-directed learning (e.g. by watching YouTube videos), and would like to have a clear understanding of who is available to support them when they need help with their data. These interview responses related to training, in the context of the fast-paced and evolving research landscape (explosion of big data[26]; movement toward open science[27], data sharing mandates[28], and multidisciplinary teams[29], suggest that a more formal program of data management training would be useful to the research community.
Another theme arising from the interviews was the challenge of collaborating on large projects and sharing data more widely. For those working on large, cross-institutional projects, sharing large datasets among collaborators could be challenging as was re-integrating data from side projects; difficulty in transferring data across platforms was also identified by Rambo as a significant barrier, particularly among clinical researchers.[11] Although not directly related to data, several participants mentioned the challenge of learning about resources and potential collaborators across the institution. When asked about data sharing, most participants responded that they were typically sharing only with their immediate collaborators. The main exception to this was individuals generating genetic or genomic data, who deposit this data in federal repositories as mandated by NIH. When given the option in the survey, 26.1 percent of participants stated they were using federal repositories to share data; however, when unprompted in the interviews, participants did not immediately identify their data deposits (done primarily for regulatory compliance) as being a form of data sharing. Although others expressed some interest in sharing their data, they either questioned the value of their current data to others, lacked knowledge of how to best share large data sets, or had not yet been asked to share their data. As one faculty member put it, “Do I share the data? Usually not, because there is no mechanism for that. The data is usually not shared because there is strict confidentiality involved.” Similarly, another researcher was asked whether one of her data sets had been submitted to the GEO database at NCBI and responded, “That one we didn’t because, first of all, it never really even occurred to us but also wasn’t NIH funded…” Overall, participants’ comments indicated that funding agencies’ expectations had the largest impact on their data sharing practices. Participants also found it difficult to find and use existing data (shared by others) that would be relevant to their own research. In particular, those working with genetic data discussed the challenge in keeping up with the data, and even the databases in which they are housed: “It’s so hard to keep up with the genetics databases now. They keep changing. They keep changing how you search them. There’s always new ones.”
Participants seemed largely satisfied with their current data storage practices, but long-term preservation and accessibility were more of a challenge, matching reports elsewhere in the literature of storage and preservation as one area of anticipated future need.[20] Most of the participants used college- or department-level network servers and felt that they were sufficiently reliable. Some, however, found these networks to be difficult to access remotely and preferred to use personal computers, USB drives, and external hard drives. For data from completed projects, participants discussed the inaccessibility of materials created through old or obsolete versions of software. As one faculty member put it, “Something older than 10 years I can’t probably even open it.” A number of the participants discussed and showed the librarians their print lab notebooks. Despite interest in migrating to electronic documentation, print lab notebooks were cited as the gold standard for documenting ethical conduct of research, easier to use when doing wet bench research, and less expensive than electronic options. One faculty member described the situation as follows: “Traditionally we have hand-written lab notebooks that are that way. We’ve tried to make electronic ones, but there are issues [intellectual property and compliance]. So there’s things called electronic lab books, but they cost a fortune!” Those working with biological samples indicated that their retention is even more important than the retention of some electronic data, because experiments cannot be duplicated exactly if samples are lost. This also has implications for the quality of metadata needed for this kind of data and other organizational strategies required to identify and locate them if researchers need to use them again in the future.
Several other overall concerns arose throughout the interviews, although they were not discussed as extensively as the themes above. Those working with particularly sensitive (e.g. from high-containment labs or the Veterans Affairs hospital) were concerned with balancing necessary security precautions with the usability of data within researchers’ workflows. Participants in resource-rich labs had fewer problems with data management overall, because they were able to hire staff members dedicated to handling the data. Some participants were interested in institution-level policies for data management, in response to funders’ pressures: “… we should have one policy at the level of the university, because that is one of the most important things for National Science Foundation right now.” Participants noted that in some cases, sharing will be mediated by the Institutional Review Board (IRB), which has a key concern in the security of human-subjects’ data. As one participant put it, “we are moving towards… having a gate-keeper, which is mostly an IRB issue. To decide who gets access to this stuff.” All participants, even those who currently have few data management challenges, are expecting to work with bigger, more complex data sets in the future. A faculty member described this expanding scope of research: “There are going to be thousands of people sequenced very soon. And that is a lot of data. So everybody’s in this mess. It’s just an onslaught of data that’s going to be meaningless until you have a way to look at it.”
Potential limitations
One limitation of this study is its low survey response rate (7 percent), which may introduce response bias. Previous web-based surveys of biomedical professionals, however, show that response rates under 20 percent are not uncommon and that emailed surveys continue to have lower response rates than mailed paper surveys.[30][31] Another potential limitation was the interview recruiting method of inviting individuals who had interacted with the library in the past. Using this convenience sampling method may have introduced volunteer bias, with those who agreed to participate doing so out of a particular interest in the subject matter. While this may not lead to a full picture of data management needs across UF researchers, it is likely that individuals who have previous experience with the library may have an expanded view of what librarians are capable of, and thus may produce more detailed and usable answers than the uninformed participant. Despite its potential limitations, this is the first study assessing the research data management needs of the CTSI-affiliated researchers. In addition, it has provided a basis for further research at our institution to identify likely solutions and to support research data management. Even if survey and interview responses are not generalizable across the entire CTSI research community, they represent real needs of the individual participants, which are still important for HSCL to address.
Discussion
The survey and interviews highlighted the variety of problems encountered by researchers when dealing with their data, both those problems that the researchers themselves identified and potential problems that can be inferred from their responses. When asked directly about what resources they needed to best manage their data, respondents prioritized both technical aspects like computing expertise, software, and storage capacity and practical aspects like better organizational systems and training. Other responses — such as individuals not annotating data, thinking all of their data should be kept forever, or relying on personal computers and CDs/DVDs for storage — indicated a broader lack of awareness of best practices in data management. Given this diversity of needs and awareness, our next steps focused on training and on creating the collaborative infrastructure to work in more detail on additional data management needs.
A commonality across the diverse information collected through the survey and interviews was an interest in training on data management, particularly for students. Thus, one of the first outcomes of the assessment was the development of a workshop and accompanying LibGuide on Research Data Management at UF. Two of the authors (RGM, HFN) developed a guide (http://guides.uflib.ufl.edu/datamanagement), which provides an overview of the types of issues involved in effective data management and links to resources to address those issues, with a particular focus on organizations and tools within our university community. We introduced the guide at presentations during Research Computing Day, hosted by UF Research Computing, which also links to the guide from its website. Subsequently, we have had a fair amount of traffic on the page with over 4,800 hits from its inception in 2012 through February 2016. At Research Computing Day, we shared some of the survey results (including those related to storage, annotation, protection, and sharing of data) as part of a conversation among attendees about next steps in supporting research data management; in this conversation, a number of attendees commented on the need for more training across UF, reinforcing our conclusions. The Research Data Management at UF guide was used to support training beginning in the fall of 2012, with the creation of the one-hour workshop “Best Practices in Research Data Management.” This session is taught within HSCL’s stand-alone workshop series — drop-in sessions that are advertised to Health Science Center students, faculty, and staff, but open to anyone at UF. The session is organized as a discussion of the principles and resources included on the LibGuide and often includes participants sharing their data management challenges and solutions, including suggestions for useful tools. We have taught 11 sessions of this workshop to a total of 42 attendees, including faculty, students, and staff from five of the six Health Science Center colleges (Medicine, Public Health and Health Professions, Dentistry, Nursing, and Pharmacy); this is moderate-to-high attendance for the HSC Library’s stand-alone workshop series. This workshop was designed primarily with graduate students in mind, but we have received feedback through the graduate programs that students are typically unwilling to attend stand-alone workshops without academic credit. At the suggestion of one attendee, a session was developed specifically for clinical research coordinators (40 attended) and was taught in collaboration with CTSI’s REDCap support staff.
Two other venues for instruction in data management have been developed at the HSCL. Since 2013, the liaison librarian who works with Ph.D. students in the College of Medicine has provided a short introduction to the topic and associated LibGuide during her library orientation for new students in the IDP (Interdisciplinary Program in Biomedical Sciences) — approximately 25-40 students per year. HSCL liaison librarians who are primarily responsible for serving professional students of the six colleges are encouraged to do the same in their orientations and course-integrated instruction. A more detailed discussion of the topic will be provided in “Data Management: Best Practices, Requirements, Resources,” a 90-minute module in the HSCL’s newly created credit-bearing course “Finding Biomedical Research Information and Communicating Science,” targeted to graduate students in the College of Medicine. This course was designed, in part, in response to the feedback mentioned above that graduate students were interested in the topics of our stand-alone workshops but unable to devote the time to sessions without academic credit. The data management module will introduce students to practical strategies for managing research data as well as to data management tools and resources available at the local and national levels, and will provide an overview of data management planning and sharing from the perspective of funding agencies. Students in the course will consider questions central to managing research data, including those addressing data collection, metadata and annotation, storage, security, and data sharing. Students will use case studies to explore data management issues with the expectation that they will be able to apply the same kinds of questions and planning to their own research data following completion of this session. The course will be offered in the fall of 2016 and will include modules on literature searching and management, research impact, compliance, and other topics of interest to graduate students.
Moving beyond the CTSI and HSCL, the other major outcome of this assessment was the formation of UF’s Data Management/Curation Task Force (DMCTF). The range of needs identified in the needs assessment were diverse and included basic needs in identifying storage venues, strategies and assistance for preserving data, organizing data and making them retrievable through the use of appropriate metadata; this was the first concrete evidence that these needs existed across the UF community — many coincided very clearly with traditional library services for collecting, organizing, providing access, and preserving resources. The central role for the library seemed clear — what was less apparent was a clear identification of what resources were currently available to researchers at UF. The Data Management/Curation Task Force was conceived as a collaborative working group representing various entities on campus with interest in the future of data management in general at the University, notably the libraries, Research Computing, and the Office of Research. This group was called to begin work in early 2013 by HSCL’s Director (author CB), who is also Associate Dean of the George A. Smathers Libraries. The DMCTF was entrusted to determine the current landscape surrounding the collection, organization, dissemination and preservation of data on campus. In addition, they were asked to identify and propose specific service areas for the library in campus level data-related activities. Finally, the libraries have traditionally had a core role in providing training to end users and the DMCTF was charged with identifying the training needs and devising a plan for meeting those needs. The goal was for the group to develop training materials and opportunities for library liaisons who are then tasked with providing training to the end user, as in the Purdue model.[4]
The DMCTF has performed a wide variety of activities in working to fulfill its charge, including the following:
- Assessing data-related needs across campus through a survey (modeled on the HSCL survey) and focus group sessions
- Customizing DMPtool (https://dmptool.org) with UF authentication and links
- Hosting a half-day event “Big Data, Little Data” targeted towards graduate students
- Offering the five-session series “Core Data Training for Reference Services” for librarians and library staff
- Developing template materials for liaison librarians and subject specialists to use when discussing data management with their users
The DMCTF’s most recent focus has been on developing “Data Management Guidelines & Best Practices to Assist in Research Data Policy Development.” As the title implies, this document presents some guidelines and best practices with a focus on use of existing institutional resources, and is designed to initiate further conversation with campus stakeholders before a more explicit data management policy development is required. To that end, it has been distributed to UF’s Research Computing Advisory Committee, Informatics Institute, Office of Research, and Faculty Senate IT Subcommittee for comments and changes.
Throughout all of its activities, the DMCTF has worked towards developing a culture of data management in the libraries and beyond. To bring new expertise, insights, and leadership to the DMCTF and dedicated effort to the libraries’ goals in data management support, the George A. Smathers Libraries have recently hired a Data Management Librarian, who started at UF in January of 2016.
Conclusion
Survey and interview results indicate that UF’s CTS researchers have diverse data management needs that are often specific to their discipline or current research project and span the data lifecycle. While these diverse needs call for a wide variety of potential solutions, HSCL and the George A. Smathers Libraries have begun addressing common campus-wide concerns through data management training, collaboration with campus IT infrastructure and research units, and creating a Data Management Librarian position. Initiating conversations with affected stakeholders and campus leadership about best practices in data management and implications for institutional policy shows the library’s proactive leadership in this area and furthers our goal to provide concrete guidance to our users.
Supplemental content
Appendix 1: University of Florida Health Science Center Library Data Survey - Appendix1_1090.pdf (684 kB)
Appendix 2: UF Health Science Center Library Data Interview Questions – Modified from University of Virginia - Appendix2_1090.pdf (694 kB)
Acknowledgements
This project has been funded in part with federal funds from the National Library of Medicine, National Institutes of Health, under Contract #HHS-N-276-2011-00004-C.
Disclosure
The author(s) report no conflict of interest.
References
- ↑ National Science Board (September 2005). "Long-Lived Digital Data Collections Enabling Research and Education in the 21st Century". National Science Foundation. pp. 89. http://www.nsf.gov/pubs/2005/nsb0540/.
- ↑ Gold, A. (2007). "Cyberinfrastructure, Data, and Libraries, Part 2: Libraries and the Data Challenge: Roles and Actions for Libraries". D-Lib Magazine 13. doi:10.1045/july20september-gold-pt2.
- ↑ Charbonneau, D.H. (2013). "Strategies for Data Management Engagement". Medical Reference Services Quarterly 32 (3): 365-74. doi:10.1080/02763869.2013.807089. PMID 23869641.
- ↑ 4.0 4.1 Garritano, J.R.; Carlson, J.R. (2009). "A Subject Librarian's Guide to Collaborating on e-Science Projects". Issues in Science and Technology Librarianship 57 (Spring 2009). doi:10.5062/F42B8VZ3.
- ↑ Heidorn, P.B. (2011). "The Emerging Role of Libraries in Data Curation and E-science". Journal of Library Administration 51 (7–8): 662–672. doi:10.1080/01930826.2011.601269.
- ↑ Rambo, N. (2009). "E-science and biomedical libraries". Journal of the Medical Library Association 97 (3): 159–161. doi:10.3163/1536-5050.97.3.001. PMC PMC2706433. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2706433.
- ↑ Reed, R.B. (2015). "Diving into Data: Planning a Research Data Management Event". Journal of eScience Librarianship 4 (1): e1071. doi:10.7191/jeslib.2015.1071.
- ↑ 8.0 8.1 8.2 Peters, C.; Vaughn, P. (2014). "Initiating Data Management Instruction to Graduate Students at the University of Houston Using the New England Collaborative Data Management Curriculum". Journal of eScience Librarianship 3 (1): e1064. doi:10.7191/jeslib.2014.1064.
- ↑ Goldman, J.; Kafel, D.; Martin, E.R. (2015). "Assessment of Data Management Services at New England Region Resource Libraries". Journal of eScience Librarianship 4 (1): e1068. doi:10.7191/jeslib.2015.1068.
- ↑ Piorun, M.E.; Kafel, D.; Leger-Hornby, T. et al. (2012). "Teaching Research Data Management: An Undergraduate/Graduate Curriculum". Journal of eScience Librarianship 1 (1): e1003. doi:10.7191/jeslib.2012.1003.
- ↑ 11.0 11.1 11.2 11.3 Rambo, Neil (22 October 2015). "Research Data Management Roles for Libraries" (PDF). http://www.sr.ithaka.org/wp-content/uploads/2015/10/SR-Issue_Brief_Research_Data_Management_1022151.pdf.
- ↑ Nelson, M.S. (2015). "Data Management Outreach to Junior Faculty Members: A Case Study". Journal of eScience Librarianship 4 (1): e1076. doi:10.7191/jeslib.2015.1076.
- ↑ Reznik-Zellen, R.C.; Adamick, J.; McGinty, S. (2012). "Tiers of Research Data Support Services". Journal of eScience Librarianship 1 (1): e1002. doi:10.7191/jeslib.2012.1002.
- ↑ Foster, N.F.; Gibbons, S., ed. (2007) (PDF). Studying Students: The Undergraduate Research Project at the University of Rochester. Chicago: Association of College and Research Libraries. pp. 90. ISBN 9780838984376. http://www.ala.org/acrl/sites/ala.org.acrl/files/content/publications/booksanddigitalresources/digital/Foster-Gibbons_cmpd.pdf.
- ↑ 15.0 15.1 Anderson, N.R.; Lee, S.; Brockenbrough, J.S. et al. (2007). "Issues in Biomedical Research Data Management and Analysis: Needs and Barriers". JAMIA 14 (4): 478–488. doi:10.1197/jamia.M2114. PMC PMC2244904. PMID 17460139. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2244904.
- ↑ 16.0 16.1 Witt, M.; Carlson, J.; Brandt, D.S.; Cragin, M.H. (2009). "Constructing Data Curation Profiles". International Journal of Digital Curation 4 (3): 93–103. doi:10.2218/ijdc.v4i3.117.
- ↑ Bardyn, T.P.; Resnick, T.; Camina, S.K. (2012). "Translational Researchers’ Perceptions of Data Management Practices and Data Curation Needs: Findings from a Focus Group in an Academic Health Sciences Library". Journal of Web Librarianship 6 (4): 274–287. doi:10.1080/19322909.2012.730375.
- ↑ Reich, M.; Shipman, J.P.; Narus, S.P. et al. (2013). "Assessing clinical researchers' information needs to create responsive portals and tools: My Research Assistant (MyRA) at the University of Utah: A case study". Journal of the Medical Library Association 101 (1): 4–11. doi:10.3163/1536-5050.101.1.002. PMC PMC3543136. PMID 23405041. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3543136.
- ↑ Guindon, A. (2014). "Research Data Management at Concordia University: A Survey of Current Practices" (PDF). Feliciter 60 (2): 15–17. http://cla.ca/wp-content/uploads/60_2.pdf.
- ↑ 20.0 20.1 Weller, T.; Monroe-Gulick, A. (2015). "Differences in the Data Practices, Challenges, and Future Needs of Graduate Students and Faculty Members". Journal of eScience Librarianship 4 (1): e1070. doi:10.7191/jeslib.2015.1070.
- ↑ Soehner, C.; Steeves, C.; Ward, J. (23 June 2010). "e-Science and data support services: a survey of ARL members". 31st Annual IATUL Conference. Purdue University. http://docs.lib.purdue.edu/iatul2010/conf/day3/1/.
- ↑ National Center for Research Resources (2009). "Clinical and Translational Science Awards: Advancing Scientific Discoveries Nationwide to Improve Health" (PDF). National Institutes of Health. pp. 37. https://ncats.nih.gov/files/CTSA-report-2006-2008.pdf.
- ↑ Guzick, D.S. (8 October 2015). "Clinical, Translational and Implementation Science: Part 1 - CTSA renewal". UFHealth. University of Florida. https://ufhealth.org/news/2015/clinical-translational-and-implementation-science-part-1-ctsa-renewal.
- ↑ Barribal, K.L.; While, A. (1994). "Collecting data using a semi-structured interview: A discussion paper". Journal of Advanced Nursing 19 (2): 328–335. doi:10.1111/j.1365-2648.1994.tb01088.x. PMID 8188965.
- ↑ Harris, P.A.; Taylor, R.; Thielke, R. et al. (2009). "Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support". Journal of Biomedical Informatics 42 (2): 377–381. doi:10.1016/j.jbi.2008.08.010.
- ↑ Anderson, J.Q.; Rainie, L. (20 July 2012). "Big Data: Experts say new forms of information analysis will help people be more nimble and adaptive, but worry over humans’ capacity to understand and use these new tools well" (PDF). Pew Internet. Pew Research Center. http://www.pewinternet.org/files/old-media/Files/Reports/2012/PIP_Future_of_Internet_2012_Big_Data.pdf.
- ↑ Morey, R.D.; Chambers, C.D.; Etchells, P.J. et al. (2016). "The Peer Reviewers' Openness Initiative: incentivizing open research practices through peer review". Royal Society Open Science 3 (1): 150547. doi:10.1098/rsos.150547.
- ↑ Burwell, S.M.; VanRoekel, S.; Park, T.; Mancini, D.J. (9 May 2013). "Open Data Policy - Managing Information as an Asset" (PDF). U.S. Office of Management and Budget. https://www.whitehouse.gov/sites/default/files/omb/memoranda/2013/m-13-13.pdf.
- ↑ Disis, M.L.; Slattery, J.T. (2010). "The Road We Must Take: Multidisciplinary Team Science". Science Translational Medicine 2 (22): 22cm9. doi:10.1126/scitranslmed.3000421.
- ↑ Hardigan, P.C.; Popovici, I.; Carvajal, M.J. (2016). "Response rate, response time, and economic costs of survey research: A randomized trial of practicing pharmacists". Research in Social & Administrative Pharmacy 12 (1): 141–148. doi:10.1016/j.sapharm.2015.07.003.
- ↑ Scott, A.; Jeon, S.-H.; Joyce, C.M. et al. (2011). "A randomised trial and economic evaluation of the effect of response mode on response rate, response bias, and item non-response in a survey of doctors". BMC Medical Research Methodology 11: 126. doi:10.1186/1471-2288-11-126.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. References were originally listed alphabetically; they were converted to the standard wiki inline format, in order of appearance.