Journal:Design of generalized search interfaces for health informatics
| Full article title | Design of generalized search interfaces for health informatics |
|---|---|
| Journal | Information |
| Author(s) | Demelo, Jonathan; Sedig, Kamran |
| Author affiliation(s) | Western University |
| Primary contact | Email: sedig at uwo dot ca |
| Editors | Almada, Marta |
| Year published | 2021 |
| Volume and issue | 12(8) |
| Article # | 317 |
| DOI | 10.3390/info12080317 |
| ISSN | 2078-2489 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.mdpi.com/2078-2489/12/8/317/htm |
| Download | https://www.mdpi.com/2078-2489/12/8/317/pdf (PDF) |
Abstract
In this paper, we investigate ontology-supported interfaces for health informatics search tasks involving large document sets. We begin by providing background on health informatics, machine learning, and ontologies. We review leading research on health informatics search tasks to help formulate high-level design criteria. We then use these criteria to examine traditional design strategies for search interfaces. To demonstrate the utility of the criteria, we apply them to the design of the ONTology-supported Search Interface (ONTSI), a demonstrative, prototype system. ONTSI allows users to plug-and-play document sets and expert-defined domain ontologies through a generalized search interface. ONTSI’s goal is to help align users’ common vocabulary with the domain-specific vocabulary of the plug-and-play document set. We describe the functioning and utility of ONTSI in health informatics search tasks through a workflow and a scenario. We conclude with a summary of ongoing evaluations, limitations, and future research.
Keywords: information search, search tasks, health informatics, interface design, ontologies, machine learning, PubMed
Introduction
Health informatics is concerned with emergent technological systems that improve the quality and availability of care, promote the sharing of knowledge, and support the performance of proactive health and wellness tasks by motivated individuals.[1] Subareas of health informatics may include medical informatics, nursing informatics, consumer informatics, cancer informatics, and pharmacy informatics, to name a few. Simply put, health informatics is concerned with harnessing technology for finding new ways to help stakeholders work with health information to be able to perform health-related tasks more effectively.
Users in the health domain are increasingly taking advantage of computer-based resources in their tasks. For instance, a 2017 Canadian survey found that 32% of respondents within their last month had used at least one mobile application for health-related tasks. Even more, those under the age of 35 are twice as likely to do so.[2] Furthermore, studies have calculated that over 58% of Americans have used tools like Google and other domain-specific tools to support their health informatics search tasks, with search being one of the most important and central tasks in most health informatics activities.[3][4]
Yet, search can be challenging, particularly for health informatics tasks that utilize large and complex document sets. For such tasks, health informatics tools may require the use of domain-specific vocabulary. Aligning with this vocabulary can be a significant challenge within health tasks, as they can involve a lexicon of intricate nomenclature, deeply layered relations, and lengthy descriptions that are misaligned with common vocabulary. For instance, one highly cited medical research paper defines the term “chromosomal instability” as “an elevated rate of chromosome mis-segregation and breakage, results in diverse chromosomal aberrations in tumor cell populations.” In this example, those unfamiliar with the defined term could find parsing its definition just as significant a challenge as the term itself.[5] Thus, when communicating across vocabularies, users may struggle to describe the requirements of their search task in a way that is understandable by health informatics tools.[6][7] To deal with this challenge, ontologies can be a valuable mediating resource in the design of user-facing interfaces of health informatics tools.[8] That is, ontologies can bridge the vocabularies of users with the vocabulary of their task and its tools. Yet, the use of ontologies in user-facing interface design is not well established. Furthermore, health informatics tools that present a generalized interface, one that can support search tasks across any number of domain vocabularies and document sets, can allow users to transfer their experience between tasks, presenting users with information-centric perspectives during their performances rather than technology-centered perspectives.[9][10] For this, there is a need to distill criteria that can guide designers during the creation of ontology-supported interfaces for health informatics search tasks involving large document sets.
The goal of this paper is to investigate the following research questions:
- What are the criteria for the structure and design of generalized ontology-supported interfaces for health informatics search tasks involving large document sets?
- If such criteria can be distilled, can they then be used to help create such interfaces?
In this paper, we examine health informatics, machine learning, and ontologies. We then review leading research on health informatics search tasks. From this analysis, we formulate criteria for the design of ontology-supported interfaces for health informatics search tasks involving large document sets. We then use these criteria to contrast the traditional design strategies for search interfaces. To demonstrate the utility of the criteria in design, we will use them to structure the design of a tool, ONTSI (ONTology-supported Search Interface). ONTSI allows users to plug-and-play their document sets and expert-defined ontology files to perform health informatics search tasks. We describe ONTSI through a functional workflow and an illustrative usage scenario. We conclude with a summary of ongoing evaluation efforts, future research, and our limitations.[11]
Background
In this section, we describe the concepts and terminology used when discussing ontology-supported interfaces for health informatics search tasks involving large document sets. We begin with background on health informatics. Next, we examine machine learning. We conclude with coverage of ontologies and their utility as a mediating resource for both human- and computer-facing use.
Health informatics
Health informatics is broadly concerned with emergent technological systems for improving the quality and availability of care, promoting the sharing of knowledge, and supporting the performance of proactive health and wellness practices by motivated individuals.[1] Initially, the need for expanded health and wellness services stemmed from rising population levels combined with the growing complexity of medical sciences. These issues made it challenging to maintain quality care within increasingly stressed medical systems.[12] Thus, a central objective for health informatics is the development of strategies to tackle large-scale problems that harm trained medical professionals’ ability to perform their tasks in a timely and effective manner. For instance, telehealth services allowed doctors to practice remote medicine, providing care to those without local medical services. Another early innovation was standardized electronic health care records (EHRs), where patient records were given standardized encodings to provide an increased ability to track, compare, manage, and share personal health information.[3] Some examples of current research directions are the push for stronger patient privacy, personalized medicine, and the expansion of healthcare into underserved regions and communities.[1][2][3][13][14]
The rising production and availability of health-related data has resulted in a growing number of data-intensive tasks within health. Both private and public entities like health industry companies, government bodies, and everyday citizens are turning to health informatics tools as they manage and activate their health data.[2] A growing number of health-related tasks involve searching document sets. During these tasks, the aim of the user is to use the information described within their document set to increase their understanding of a topic or concept. For example, a search task could be a practitioner searching the EHRs of their patients, a member of the general public using public materials for their general health concerns, or a researcher performing a literature review.[12][15][16] In general, a search task involves the generation of a query based on an information-seeking objective. The computation systems of these tools then use this query to map and extract relevant documents out from the document set.[15] Powerful technologies like machine learning are increasingly being integrated within tools to help perform rapid and automated computation on document sets.[4] Yet, when taking advantage of these technologies, designers must be mindful of human factors when generating the user-facing interfaces of their tools, as a task cannot be performed effectively without direction from an empowered user.[12]
Machine learning
Machine learning techniques are increasingly being utilized to tackle analytic problems once considered too complex to solve in an effective and timely manner.[16] Yet, recent analysis[17][18][19] on the human factors in machine learning environments have found that the current design strategies continually limit users’ ability to take part in the analytic process. More so, it has produced a generation of machine learning-integrated tools that are failing to provide users a complete understanding on how computational systems of their tools arrive at their results. This has significantly reduced users’ control and lowered the ability to achieve task objectives. In response, there is a growing desire to promote the “human-in-the-loop,” bringing the benefits of human reasoning back to the forefront of the design process.[20][21][22]
When considering the interaction loop of a machine learning-integrated tool, Sacha et al.[23] present a five-stage conceptual framework: producing and accessing data, preparing data for tool use, selecting a machine learning model, visualizing computation in the tool interface, and users applying analytic reasoning to validate and direct further use. Assessing this framework, a machine learning-integrated search tool must provide users with a functional workflow where:
- users communicate their task requirements as a query;
- users ask their tool to apply that query as input within its computational system;
- the tool performs its computation, mapping the features against the document set;
- the tool represents the results of the computation in its interface;
- users assess whether they are or are not satisfied with the results; and
- users restart the interaction loop with adjustments or conclude their use of the tool.
Thus, a primary responsibility for users within machine learning environments is the need to assess how well the results of machine learning have aligned with their task objectives. A systematic review by Amershi et al.[24] suggests six considerations for the user’s role in arbitrating machine learning performance:
- Users are people, not oracles (i.e., they should not be expected to repeatedly answer whether a model is right or wrong).
- People tend to give more positive than negative feedback.
- People need a demonstration of how machine learning should behave.
- People naturally want to provide more than just data labels.
- People value transparency.
- Transparency can help people provide better labels.
Ontologies
In search tasks involving large document sets, many challenges can arise that reduce performance quality, harm user satisfaction, and increase the time for task completion.[17][18][19] Often, these challenges result from misalignment between the vocabularies used by the document sets, storage maintainers, interface designers, and users. For instance, Qing et al.[25] outline the difficulties faced when translating between common and domain vocabularies in health tasks. They describe a study that found that up to 50% of health expressions by consumers were not represented by public health vocabularies.[25]
Within the pipeline of a search task, both the human and computational system can only perform optimally if communication is strong.[26] Ontologies are representational artifacts that reflect the entities, relations, and structures of its domain. Ontologies are of three types: a philosophical ontology for describing and structuring reality, a domain ontology for structuring the entities and relations of a knowledge base, and a top-level ontology for interfacing between different domain ontologies.[26] Ontologies provide the flexibility, extensibility, generality, and expressiveness necessary to bridge the gap when mapping domain knowledge for effective computer-facing and human-facing use.[8] For this purpose, ontologies are increasingly being used within tools to help users perform their challenging search tasks.[27][28][29][30][31]
When creating an ontology, experts construct a network of entities and relations, which together yield various structures.[32][33] Ontology entities reflect the objects of the domain, like a phenotype in a medical abnormality ontology, a processor in a computer architecture ontology, or a precedent in a legal ontology.[34] In some ontologies, like the top-level ontology, Basic Formal Ontology, designers go as far as denoting qualities such as materiality, object composition, and spatial qualities in reality.[26] Ontology entities are encoded with information about their role in the vocabulary, definitions, descriptions, and contexts, as well as metadata that can inform the performance of future ontology engineering tasks.
Ontology relations are the links between ontology entities that express the quality of interaction between them and the domain as a whole.[35] When assessing ontology relations, Arp et al.[26] distinguish relations under the categories of universal–universal (dog “is_a” animal), particular–universal (this dog “instance_of” dog), and particular–particular (this dog “continuant_parts” of this dog grouping). Domain ontology relations are realized through unique interoperability between ontology entities. For instance, an animal ontology may have an ontology entity reflecting the concept of a “human,” which may have the ontology relations “domesticates/is domesticated by” between it and the “dog” ontology entity.
After defining the entities, relations, and other features of an ontology, experts record their work in ontology files of standardized data formats like RDF, OWL, and OBO. These ontology files are then distributed amongst users. They can then be integrated into the computational and human-facing systems of tools for use during tasks. Some examples of current ontology use are information extraction on unstructured text, behavior modeling of intellectual agents, and an increasing number of human-facing visualization tasks such as decision support systems within critical care environments.[20][21][22]
Methods
In this section, we describe the methods used for criteria formulation. We begin with a review of literature for health informatics search tasks. Based on the insights gained from this review, we distill a set of criteria. We then use these criteria to contrast traditional design strategies for interfaces of search tasks.
Task review
Here, we review some research on interfaces for health informatics search tasks. We used Google Scholar, IEEE Xplore, and PubMed to conduct an exhaustive search of articles and reviews published between 2015 and 2021. We have divided our findings into three sections. First, we explore research on health data, information management, and information-centric interfaces. This is followed by research discussing the types of search tasks and their use in structuring the design of interfaces for health informatics. Finally, we investigate the requirements for aligning vocabularies for health informatics search tasks.
Health data, information management, and information-centric interfaces
Health data is constantly generated, highlighted by reports that within just a year the U.S. healthcare system created 150 new exabytes of data.[36] Yet, the information that is expressed by this data, such as personal medical records, research publications, and consumer health media, is not useful unless it can be effectively understood and utilized by users. As such, it is critical to examine the challenges facing users when performing their tasks, and through this establish novel strategies for supporting the activation of health data.
Fang et al.[10] explore the pressing challenges for accessing health data under the four categories: volume, variety, velocity, and veracity. They find that the volume of health care data creates challenges in the management of data sources and stores. They describe that existing strategies are struggling, and that novel designs should be established for scaling data services. They explain that a variety of challenges come with the management of data characteristics, ranging from unstructured datapoints generated from sources like sensors, to structured data entities like research papers and medical documents. For this, they state that designers should concentrate on aligning with the characteristics of the information being encountered. Next, they explore the challenges of velocity, which involves the rate at which users require their data to move from source to activation within their task. They highlight novel research in the networking and data management space. Finally, they explore veracity challenges, such as the assessment and validation of data quality and the quality of information that the data may produce.
Gibson et al.[9] provide a review of the evolving fields of health information management and informatics. They review the topics of data capture, digital e-record systems, aggregate health management, healthcare funding models, data-oriented evidence-based medicine, consumer health applications, health governance, personal health access, and genomic personalized medicine. Similar to Fang et al.[10], they note that the predominant work for health informatics should be concerned with presenting users with information-centric perspectives during their performances rather than technology-centered perspectives. More so, they describe that users in healthcare “must often navigate and understand complex clinical workflows to effectively … capture, store, or exchange information.” In other words, task workflows are already complex; therefore, effective interfaces should promote information encounters that help users perform better, rather than engage in unrelated technical details.
From the above research, we distill the criterion: Designs should maintain an information-centric interface that is flexible with respect to the dynamic requirements of search tasks like veracity of data sources, variety of data types, and evolving needs of users for health informatics.
Search tasks and structuring the design of interfaces for health informatics
Russell-Rose et al.[37] describe professional health workplace tasks. They find that the most prevalent types of search tasks are literature reviews for overviewing a topic, scoping reviews for rapidly inspecting the possible relevance of an information source, rapid evidence reviews for appraising the overall quality of a scoping review, and, finally, systematic reviews for exploring a topic in a robust manner.
During search tasks, users often lack the ability to perceive how their query decisions impact, relate, and interact with the document set. This is an important consideration for users who might want to adjust a query to better align with their information-seeking objectives. A further study by Russell-Rose et al.[38] analyzes search strategies performed by healthcare professionals. They find that a large majority of participants have a general desire to utilize advanced search functionalities when available. This suggests that users are not hesitant to take advantage of resources that they believe help optimize their task performance. Huurdeman[39] outlines that for this, a good course of action is to leverage query corrections, autocomplete, and suggestions. Yet, they find that such additions can be harmful if those features do not provide appropriate domain context. That is, resources must allow users to be contextually aware of how their query aligns with the contents of document sets, as well as the conditions of computational technologies used by interfaces.
In the same research, Huurdeman[39] investigates complex tasks involving information search and information-seeking models when using multistage search systems. In this research, they explore requirements that designers must account for when supporting users. Challenging search tasks require users to learn about the searched domain, understand how their objectives align, and formulate their objectives into a way that can be used by their tool. In other words, query building requires users to be domain cognizant, as they must communicate information-seeking objectives in a way that is understood by the tool, yet also aligns with the information found within the document sets. Thus, a health informatics tool that supports search tasks should provide the opportunity for understanding the domain of the document set being explored.
Zahabi et al.[40] describe a set of nine requirements for designers when considering how to design usable interfaces for health informatics search tasks, summarized as:
- Naturalness: The workflow of the system must present a natural task progression.
- Consistency: The parts of the system should present similar functional language.
- Prevent errors: Be proactive in the prevention of potential errors.
- Minimizing cognitive load: Align cognitive load to the requirements of the task.
- Efficient interaction: Be efficient in the number of steps to complete a task.
- Forgiveness and feedback: Supply proper and prompt feedback opportunities.
- Effective use of language: Promote clear and understandable communication.
- Effective information presentation: Align with information characteristics.
- Customizability/flexibility: The system should remain flexible to the task requirements.
Additional research by Dudley et al.[41] reviews user interface design for machine learning environments. They provide a set of principles that can be used by designers:
- Make task goals and constraints explicit.
- Support user understanding of model uncertainty and confidence.
- Capture intent rather than input.
- Provide effective data representations.
- Exploit interactivity and promote rich interactions.
- Engage the user.
From this research, we can distill two criteria. First, designs should provide interaction loops that promote prompt and effective feedback opportunities for the user. Second, designs should provide representations that are natural and consistent to the requirements of the information source, the user, and the task.
Aligning vocabularies for health informatics search tasks
When considering interfaces for health informatics search tasks, a major challenge for users is the need to overcome problem formulation deficiencies when encountering unfamiliar domains. This is because, according to Harvey et al.[42], users have been found to consistently suffer from four major issues during the performance of search tasks:
- difficulty understanding the domain being searched;
- an inability to apply their domain expertise;
- lacking the capacity to formulate an effective search query within the interface that accurately reflects their information-seeking objective; and
- deficient understanding of how to assess results produced by search, to decide whether the search has or has not satisfied their objective.
Harvey et al.[42] show that in domains with complex vocabularies, such as health and medicine, the disparity of potential users’ prior knowledge is extreme. They find that non-expert users routinely do not possess enough domain knowledge to address their information-seeking needs. This can cause significant issues during query formulation. As a result, non-expert users must first step away from their tool to learn specialized vocabulary before they can begin query building. Both Soldaini et al.[43] and Anderson and Wischgoll[44] describe that this issue can still affect even experts. This is because experts often must make assumptions when attuning to their tool.
There is growing research targeting the generation and application of mediation resources to help reduce the communication gap while using health informatics tools. Zeng et al.[25] investigate the development of consumer health vocabularies for reducing the discourse gap between lay people and medical information document sets. Furthermore, Soldaini et al.[45] explore the use of novel query computation strategies to improve the quality of medical literature retrieval during search tasks. In their quantitative study, they contrast models generated using combinations of algorithms, vocabularies, and feature weights, assessing the computational performance of different query reformulation techniques. The results of their study suggest “greatly improved retrieval performance” when utilizing combined machine learning and bridged vocabularies. More so, they provide insight regarding the quality of options that can support computational systems for health informatics search tasks.
From the above research, we can distill two criteria. First, designs should provide interactions that allow users to efficiently prepare, perform, assess, and adjust their machine learning to align with information-seeking objectives of search tasks. Second, designs should provide mediation opportunities that assist users in communicating information-seeking objectives into the domain-specific vocabulary of the document set.
High-level criteria
In Table 1, we provide five criteria based on the above review.
| ||||||||||||||
Analysis of traditional interface strategies for health informatics search tasks
We now assess the traditional design strategies for interfaces for health informatics search tasks. Wilson’s comprehensive Search User Interface Design[46] provides a complete survey of the history and current state of search interfaces. Based on their survey, and in particular their discussion of input and control features within the modern search user interfaces, two base strategies and one extension strategy for search interfaces are realized: “structured” interfaces, “unstructured” interfaces, and, in extension, “query expansion” interfaces. Table 2 provides a summary of how the above criteria align with each interface strategy.
| ||||||||||||||||||||||||||||
Structured interface strategy
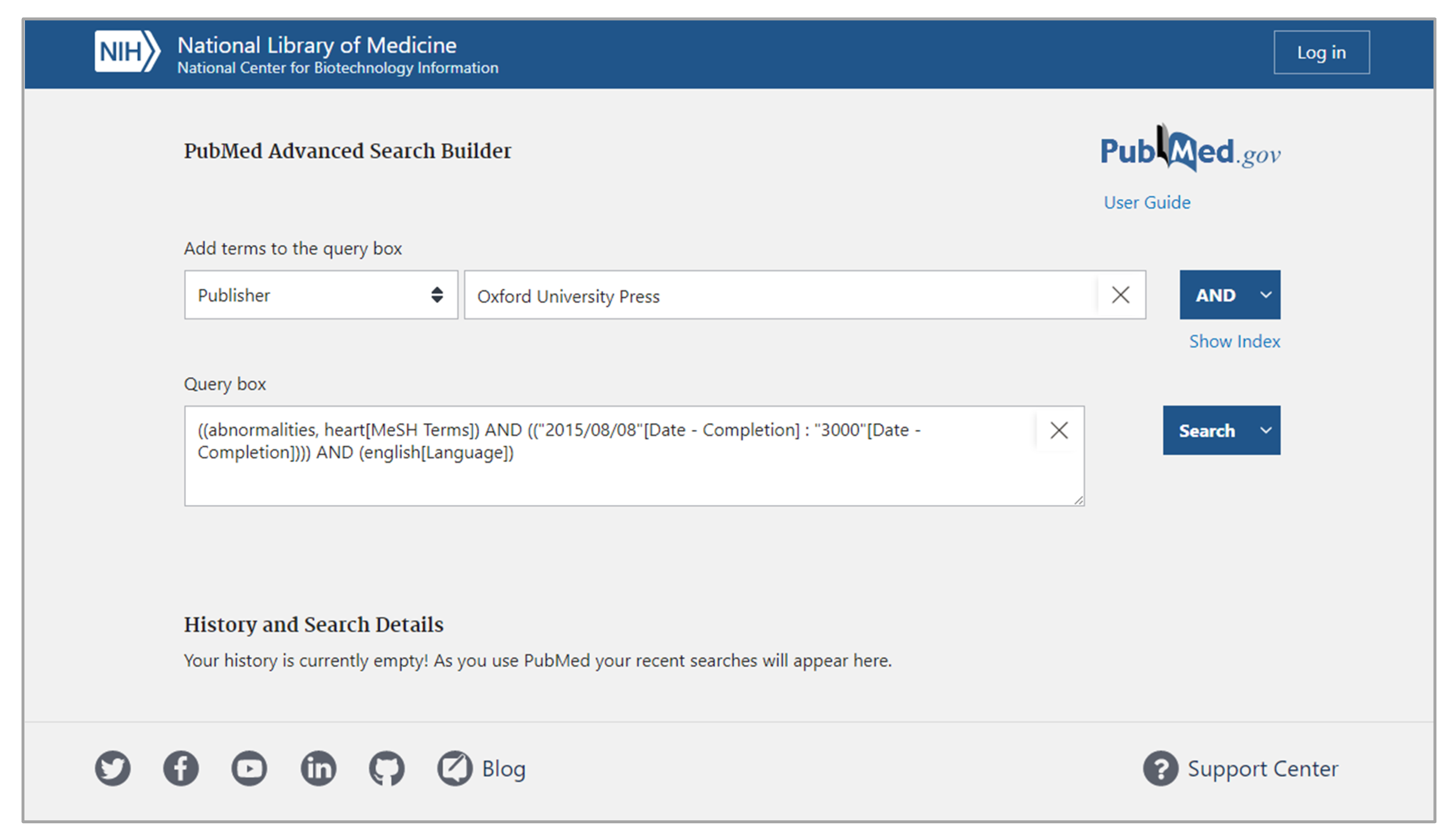
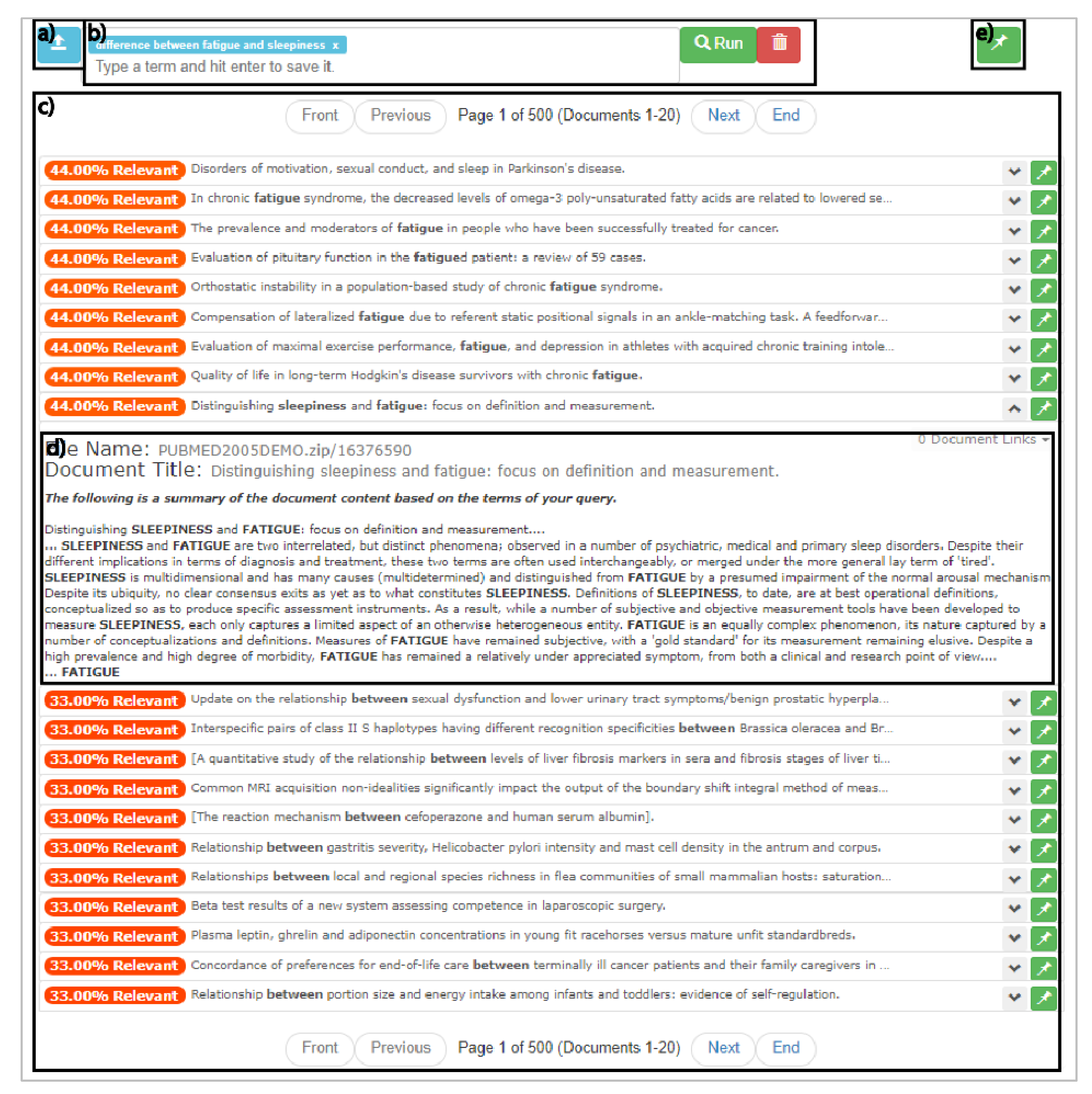
The structured interface strategy creates designs that regulate input during query building. This is achieved by maintaining heavily restricted input control profiles. Designers who implement the structured interface strategy into their interfaces presuppose a search task with specific expectations for input, bounding queries to a limited input profile. One common bounding technique is to constrain query lengths and limiting query content to a controlled set of terms.[47] This restricted scope is considered the sole acceptable input profile, and thus it allows designers to generate interfaces that limit the possible range of inputs and restrict all inputs that fall outside of that range. Designers typically achieve this by using interface elements like dropdowns, checkboxes, and radio buttons instead of elements like text boxes with free typing. For example, Figure 1 depicts the PubMed Advance Search Builder, which implements the structured interface strategy in its design. This interface requires users to select specific query term types from a restricted list, which then guides user input.[48]
|
Since input control is restricted, a strength of the structured interface strategy is that designers can use information characteristics to prescribe the full range of query formulations. This allows for the use of representational and computational designs that optimize for the expected characteristics of the restricted input profiles, per DC2. This strategy provides a designer-friendly environment that is hardened against unwanted queries, which, if effectively communicated in the design of result representations, could allow for alignment with DC3 and DC4. Yet, it can be challenging to designers to use structured interface strategies in a generalized setting. This is because when a document set is swapped, hardened approaches may not align with the information characteristics of the new document set. This negatively affects the flexibility of the interface, and in turn alignment with DC1. A potential weakness of the structured interface strategy is that it requires users to possess expertise on both the controlled vocabulary of the interface as well as the vocabulary of the document set being searched. If this is not known, user experience can suffer, drastically affecting alignment with DC5. Within the context of health informatics, such weaknesses reduce the users’ ability to effectively perform search tasks. This is because the controlled vocabularies within the health and medical domains demand significant expertise and result in numerous points of failure during the query formation process.[49][50]
Unstructured interface strategy
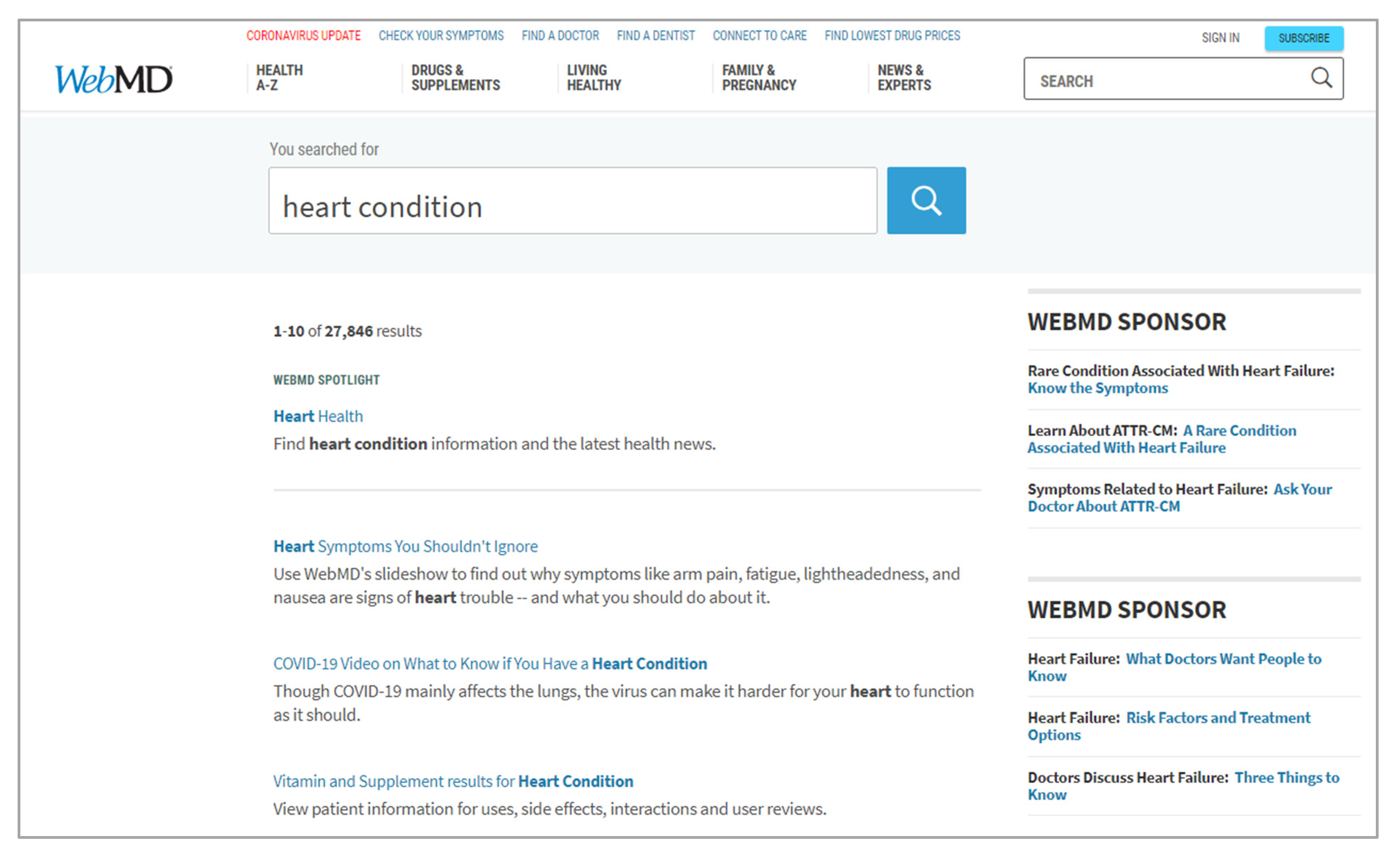
The unstructured interface strategy creates designs that provide limited input regulation. Unlike the structured interface strategy, it provides an open input control that accepts most input profiles during query building. Designers who implement the unstructured interface strategy do so without presupposing particular input, only accounting for general user error. That is, this input can originate from anywhere, such as common vocabulary, rather than from a pre-determined set of terms provided by the designer. Often, this input is directed to a single interface element. Implementations of the unstructured interface strategy typically present a text box that allows users to freely type their own text into the interface. These implementations will perform some input processing prior to use; however, the presentation of this processing to users is usually limited to correcting typographical errors rather than semantic ones. For example, the interface of Google aligns with the unstructured interface strategy, presenting users with an open, text-box input control without domain-specific assumptions or requirements. Of course, Google’s computational systems use extensive processing between receiving input from users and presenting the results of computation back to users.[51] Yet, users themselves are not informed of how their results came to be, even after changing to Google Instant.[52] Another example of an implementation of the unstructured interface strategy is WebMD’s search interface. This interface processes a free-text input with basic sanitization techniques before generating features for its search engine system, as depicted in Figure 2.
|
A strength of the unstructured interface strategy is that it supports the use of any vocabulary during query building, allowing for the natural activation of common vocabulary during task performance, in alignment with DC4. Additionally, this removes the requirement for users to possess input expertise and control profiles that typically come with a structured interface strategy, per DC1 and DC4. Designers can still implement prompt and effective feedback during task performance, thereby supporting DC2. If the constraints, processes, and results of their task performance are effectively communicated in result representations, DC3 and DC4 can be well supported. However, by allowing for the direct use of common vocabulary in lieu of a presupposed controlled vocabulary, the unstructured interface strategy suffers where the structured interface strategy excels. That is, poor implementations of the unstructured interface strategy can produce interfaces that do not provide mediation for users to translate their common vocabulary into the domain-specific vocabulary. In doing so, users are not being helped in understanding how their query building has impacted their search performance. For example, these poorly implemented interfaces may take input literally and bring users directly to a result page without providing context as to how the results were found, negatively affecting DC1 and DC5. This potential for promoting weak alignment between user and information source can lead to a significant drop in the quality of search performance. This can be an especially important requirement to address for health informatics interfaces, as it has been found that users routinely struggle to craft effective query terms during their health-related search tasks.[53]
Query expansion interface strategy
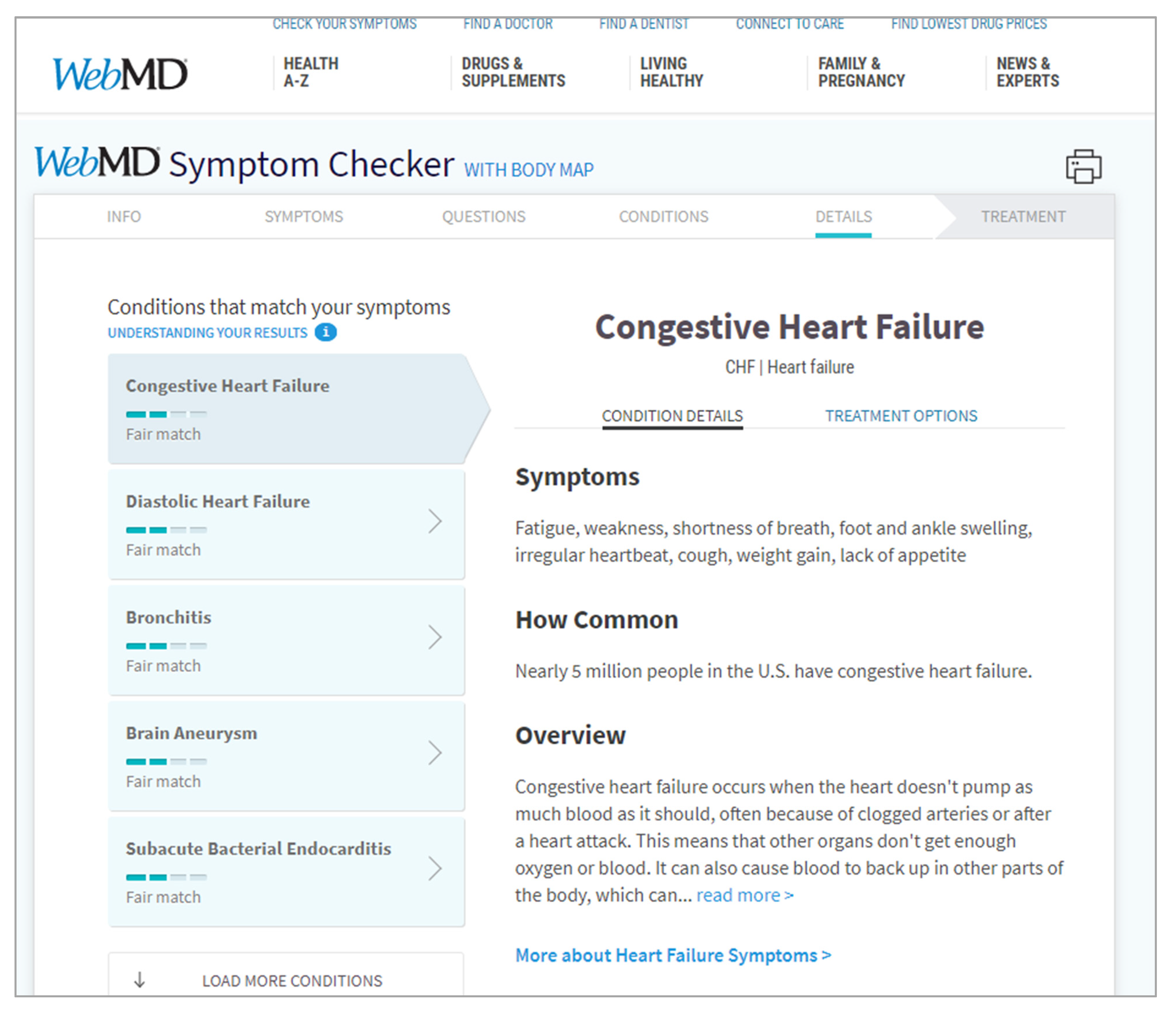
The query expansion interface strategy is an extension of both the structured and unstructured interface strategies. That is, this strategy expands by adding mediation opportunities to bridge the vocabulary of the user with the vocabulary of the document set both within the representational as well as the computational systems.[54] These mediation opportunities are typically implemented within two parts of the interaction loop. The first is during input, where mediating opportunities present during query building. Often, these mediation opportunities come as cues that suggest to users how their common vocabulary could align with the vocabulary of the domain, and visa-versa. An example of an implementation of the structured-like query expansion interface strategy is WebMD’s Symptom Checker, shown in Figure 3. This example interface goes through a series of controlled stages of query building that are structured by numerous opportunities for mediation. The second is during the processing prior to document set mapping. Like other strategies, a system can apply natural language processing techniques to the input, where the text string provided as input is tokenized into its parts. From this, the system sanitizes token parts to remove trivial tokens like the stop words “the,” “a,” and “an,” and any remaining tokens are then inserted as features in search engine systems. In more complex systems, additional sanitization techniques can be used.[55] Yet instead of immediately inserting the remaining tokens as features into the computational systems, the query expansion interface strategy builds upon the input profile by injecting insight provided by mediating resources, such as related terms, synonyms, and other expansion opportunities.[56] In other words, these systems utilize mediating resources to computationally expand the query. Some examples of mediating resources are knowledge bases like WordNet and Wikipedia, and ontologies like The Human Phenotype Ontology.[11][54][57]
|
A strength of the combined approach of the query expansion interface strategy is its strong efforts to eliminate the weaknesses associated with the structured and unstructured interface strategies while still maintaining their strengths. That is, by allowing the continued use of common vocabulary during the process of query building, users can have higher confidence about what the interface is asking of them, and what they are telling the interface to do, helping with DC4 and DC5. Furthermore, by integrating the use of mediating resources like ontologies, designers can demonstrate to users the quality of their query building and how their vocabulary decisions affect the performance of their search tasks, supporting DC2 and DC3.[58] Yet, with the added complexities of query expansion, computational systems may be required to perform more work before arriving at a final set of search results. Therefore, designers of systems taking advantage of query expansion should consider the impact on performance and responsiveness and counteract them to maintain alignment with DC2. For the query expansion interface strategy to be successful, designers must clearly communicate to users how exactly their query building has affected their search. If this communication is not provided, it can leave users confused regarding how their decisions have affected their search and can make it challenging for them to assess task performance, negatively affecting DC2. Such limitations may not provide optimal alignment in communication between the system, the user, and the information resource.[53]] That is, if a selected mediating resource does not provide an effective mapping between vocabularies, then query expansion can weaken the quality of search tasks. To address this challenge, designers can utilize user-supplied ontologies, as per DC1. This provides users the freedom to select mediating resources that they believe can best support their task performance, rather than being restricted to a tool-provided mediating resource. A user study by Jay et al.[59] compares users as they perform the same task set using two interfaces, one with a structured multiple variable input profile, the other with an unstructured single variable input profile. In this study, they find that users felt their needs and expectations were better fulfilled using the single-input profile, performing their tasks quicker, with more ease of use and learnability, and with a higher appraisal of results. Designers must carefully select how they activate query expansion such that it addresses the needs of the task, the information, and the user.
Results
In this section we describe ONTSI, a generalized ontology-supported interface for health informatics search tasks involving large document sets created using the above-discussed criteria. We outline how the criteria were used to structure ONTSI’s design. We then discuss the technical scope of ONTSI, concluding with ONTSI’s functional workflow.
Design scope
Table 3 highlights the role of each criterion in the design of ONTSI.
| ||||||||||||||
Technical scope
ONTSI is developed as a web-based tool that provides a generalized, plug-and-play support of user-supplied ontology files and document sets. That is, ONTSI allows for the uploading of ontology files, either individually or within a .zip compressed file, as well as any compressed document set in the .zip format. ONTSI then processes and indexes their contents for use within the interface. ONTSI’s front end uses the latest HTML5, CSS, and JavaScript technologies, allowing for cross-browser (i.e., Firefox, Chrome, Opera) and cross-platform support. The D3.js JavaScript library is used to create the visualization and interaction experiences found throughout the front end of ONTSI.[61] ONTSI’s back-end technology is developed using a custom Python-based computational server that maintains data transfer and machine learning APIs, and with the use of Apache’s Solr system as the search indexer and engine.[60] The current ONTSI system maintains support for the live uploading of well-formed ontologies in the Ontology Web Language (OWL) format.
Functional workflow of ONTSI
ONTSI encompasses several subsystems and subviews within its workflow. Recalling the workflow description of a machine learning-integrated search tool, ONTSI allows:
- users to communicate their task requirements as a query within its Upload and Search subview;
- users to ask their tool to apply that query as input within its computational system within its Search Subview;
- the tool to perform its computation, mapping the features against the document set within its ONTSI server and Solr server;
- the tool to represent the results of the computation in its interface within its Result List and Result Item subviews;
- users to assess whether they are or are not satisfied with the results within its Result List, Result Item, and Saved List subview; and
- users to restart the interaction loop with adjustments within the Upload and Search subviews or conclude their use of the tool.
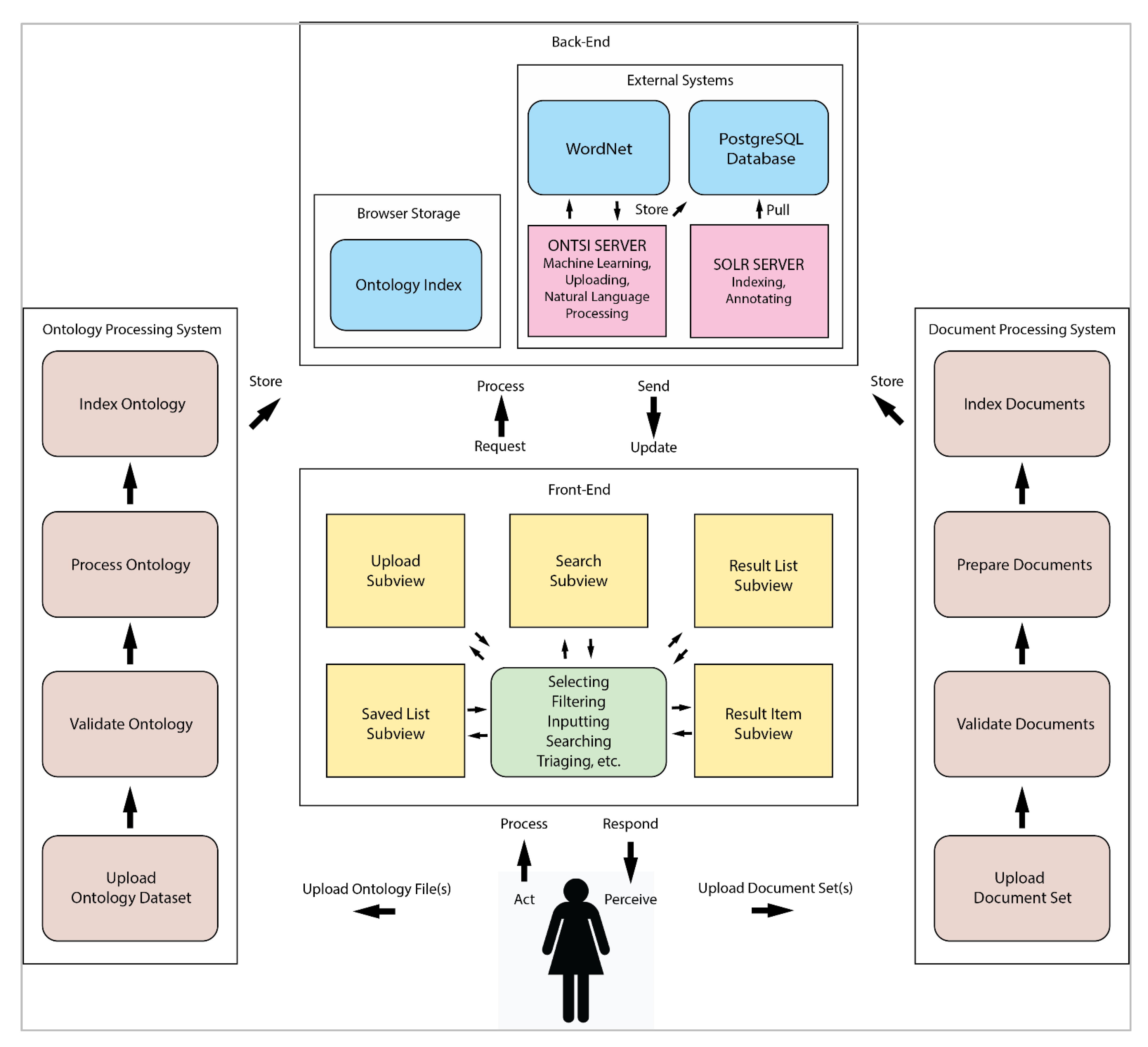
The overall functional workflow of ONTSI and its parts are described now, as depicted in Figure 4.
|
Front-end subviews
ONTSI consists of a series of interconnected subviews, shown in Figure 4 (above) and Figure 5. We will now describe the functional workflow of each subview.
|
Upload subview
The Upload subview supports the plug-and-play of user-supplied ontology files and document sets. This subview can be found at the top left of ONTSI (Figure 5a). When clicked, the upload button opens a file selection window. The window limits uploading to valid ontology files under the OWL ontology format and the .zip compression format. When a compressed file is uploaded, it is inspected for OWL files. This allows the upload system to not only take in individual OWL ontology files, but also sets of OWL files that are combined in a compressed format. Ontology file contents are put through a custom OWL to JSON processor, and then indexed into a local storage system within the browser memory. If it is a document set, it is transferred to the back end ONTSI server. Once at least one ontology file and one document set are uploaded, the Search subview and the system become active.
Search subview
The Search subview facilitates query building using an ontology-supported unstructured-like query expansion strategy. Three points of interaction are maintained: Query Input, the Run button, and the Clear button. The Search subview is located to the right of the Upload subview at the top center (Figure 5b) and becomes available for interaction after the requirements of the Upload system are fulfilled.
Query Input is a text input box. As text is typed, ONTSI cross-references that text against the uploaded ontological content for mediation opportunities. If found, those mediations are provided within an expanding dropdown. When a user values a suggested mediation, it can be selected and locked in as a query term. If none are desired, they can be ignored. When a user is satisfied with their own typed text, it can also be added. Each query term is depicted with the text of the term and a removal interaction, represented by a trailing “x” button. If multiple terms require removal, this can be done either with individual removal actions, repeated backspacing actions from the keyboard, or the red “trash can” button, which clears all query terms.
When at least one query term has been entered, the green “Run” button becomes active. This initiates the performance of computation on the uploaded document set using the ontology file for query expansion. Query terms are collected and sent to the back end ONTSI server system. The Result List subview updates when the computation is complete.
Result list subview
The Result List subview provides a paged listing the search results. The Result List subview is found directly under the Search, Upload, and Saved List subviews (Figure 5c).
Once a search is performed, the Result List subview changes from an informational alert to the results of a search. The list itself is bounded above and below by buttons and text that describe and support paging interactions. Specifically, the buttons and text describe information about the current page position, the number of pages used to divide the document set, and the number of documents in the current page, and allow for various navigation interactions on the pages.
The search results are sorted by their relevance calculation generated during clustering, such that the results assigned to document clusters that have the highest predicted relevance rating are prioritized. Then, the list is paged. Instantaneous navigation between pages is provided. Color-coded relevance ratings accompany each document, ranging from best to worst within a green–red color spectrum. Each result represents the document title with annotations highlighting terms or phrases that are believed to align with the provided query terms. A button is also provided that allows the user to access additional document content and open the document for deeper inspection. Finally, each result has a “pin” button, which allows for the saving of documents for future use within the Saved List subview.
Result item subview
The Result Item subview provides document-level information. This allows users to rapidly assess the content of individual documents during their search task. When a user selects a document within the Result List subview, ONTSI will request the full document content of that result from the Solr server using its HTTP-based API. Query terms are then used by annotation services within the Solr server to wrap HTML-based annotation tags into the document content, which is then returned to the Result Item subview. When a document is selected for inspection, the Result Item subview expands that document in place within the Result List subview, pushing down trailing items (Figure 5d).
The content of the selected document is represented in the following order: the file name of the document within the uploaded document set, the full document title, and a summarized version of the document content. The summarized version of the document content restricts the document to the passages of content that surround or have associations with the query terms provided during query building. Terms are highlighted through capitalization and with bolded font. In addition, the Result Item subview provides a dropdown at the top right, which collects all web links found within the document content for quick access. Any number of documents can be opened within the Result Item subview for comparison.
Saved list subview
Each result within the Result Item subview includes a green “pin” button, which saves documents for future reference. ONTSI collects these saved documents within the Saved List subview. The Saved List subview can be accessed at the top right of ONTSI’s overall view, directly to the right of the Search subview. There, a green “pin” Saved List can be found that allows us to request ONTSI to open the Saved List modal (Figure 5e). Upon request, the Saved List modal displays saved documents. Here, documents can be recalled, removed from the list, or copied for external use.
Back-end systems
ONTSI consists of two back-end systems that support the various front-end subviews and their controlling logic: the ONTSI server and the Solr server. Through their use, heavy computation is moved away from the browser and into dedicated computational systems. This allows for a reduction in computational overhead within the browser to improve response times and allows ONTSI to access computational technology that is not readily supported in the browser.
ONTSI server
The ONTSI server is created using the Python-based Flask framework. It exposes an application programming interface (API) supporting communication between the various systems of ONTSI. The API satisfies two major roles: preparing the uploaded document set for indexing within the Solr server and handling machine learning requests for search tasks.
When a document set has been signaled for upload within the Upload subview, it is packaged and sent through the API of the ONTSI server. Incoming document sets are assessed and provided a suitable decompression algorithm. Next, for each document within the document set, the ONTSI server assesses the encoding of that file (e.g., UTF-8, UTF-16, PDF, etc.). Based on this assessment, a suitable transcription algorithm is applied to that document. The indexing process for the Solr server is a pull interaction, so documents are stored in a static location from which they can be pulled. Therefore, the documents are sanitized, packaged, and then inserted into a temporary PostgreSQL database. The ONTSI server then requests the Solr server to begin indexing the new document set.
When a search task is initiated, the request is sent to the API of the ONTSI server. There, requests are read for settings like the clustering algorithm, the document set being searched, and query specifications. The ONTSI server then prepares the machine learning environment. Next, ONTSI performs query expansion. This involves a set of natural language preprocessing steps on the query and its individual query items, such as tokenization and the application of stop word limiters. Then, each query item is examined against the provided ontology file for mediating opportunities alongside a complete synonym ring analysis on each query item using WordNet. The original query terms and their associated ontology and synonym terms are then packaged together. These packages are then applied during the performance of unsupervised K-means clustering computation from SciKit-Learn, a third-party machine learning suite. The computed weighting characteristics of clusters are then propagated back as a package of clusters and their associated documents for the ONTSI front end for use within its various subviews. We include a pseudocode representation of these steps in Figure 6.
|
Solr server
ONTSI uses Solr, a third-party document indexing software developed by The Apache Software Foundation. Solr is a scalable indexing system that provides a valuable array of features like a REST-like API supporting many HTTP-based communication interfaces. Solr also provides and a wide range of customizable settings and schemas that supports any number of storing, searching, filtering, analysis, optimization, and monitoring tasks. For a more information regarding Solr and its various permutations, seek out their official website and documentation.[60]
A cloud-based permutation of the Solr server is used to handle the indexing and serving of uploaded document sets. Indexing occurs when a request is made to the Solr server from the ONTSI server. The Solr server schema will seek out the location of the temporary PostgreSQL database hosted by the ONTSI server, extract all new documents not already indexed, and apply a processing schema on those documents for indexing. Then, signals are sent out to the relevant ONTSI systems. Solr also handles serving requests when ONTSI requires document content, either at the metadata level when loading the Result List subview, or full, annotated document content in the Result Item subview. Requests are communicated to the Solr server through its HTTP-based API. The Solr server then handles the request, packages the results under the conditions specified in the request, and returns its response.
Usage scenario
In this section, we provide a health informatics search task scenario using ONTSI. We begin with a description of the user profile, as well as the ontology file and document set in the usage scenario. We then present the usage scenario.
User profile
The user profile we select here is that of a health stakeholder, a researcher within a professional workplace setting performing a scoping review as an information-seeking objective. A scoping review is concerned with establishing an initial idea of the amount of information on a topic within a document set.[37] The user has a general level of knowledge, typical of other health stakeholders. For instance, the user understands and can communicate phenotypic abnormalities like a broken leg, light-headedness, or loss of vision. The user understands how to perform typical actions on the interface like clicking, typing, and saving, but does not possess knowledge of the technical concerns typical of backend computational technologies.
The objective of the user is to learn whether there are any documents within a document set that are relevant to a research question. Let us assume the user’s research question is, “How does chromosomal instability drive tumor progression?” We selected this question from recently published materials on topical examples within the health domain, using “The 150 most important questions in cancer research and clinical oncology series,” published in 2017 by the Chinese Journal of Cancer.[5]
Ontology file and document set
ONTSI requires the user to upload an ontology file and a document set. We used the Human Phenotype Ontology (HPO) in the usage scenario. We selected HPO because of its high complexity resulting from its exhaustive and expert-defined domain coverage of terms and their relationships. HPO is a controlled and standardized vocabulary encoding human disease and phenotypic abnormalities. It also includes annotations in bioinformatics, biochemistry, and human genetics. HPO is an active ontology, consisting not only of over 11,000 terms, but also over 110,000 disease annotations.[62] An example of an HPO term is “blindness,” which possesses a superclass of “visual impairment,” a subclass of “congenital blindness,” and is annotated to be associated with a variety of diseases, such as a variant of colorblindness termed Achromatopsia 2.[63] Each HPO term describes attributes such as names, conceptual definitions, ontology indexing, term synonyms, class relationships, logical definitions, and expert commentary, to name a few. For additional details on the Human Phenotype Ontology, see Köhler et al. (2019)[11] and Köhler et al. (2021).[57]
The National Library of Medicine’s PubMed is selected as the document set within the usage scenario. PubMed is chosen because of its prominence within the health domain, maintaining more than 30 million citations used within a wide scope of literature and active research endeavors. Data availability limits this usage scenario to a subset of PubMed representing 10,000 document entries. These entries maintain the document title, abstract, and various metadata like authors, published date, and keywords.[48]
Usage scenario
he user loads ONTSI, finding it in its initial state, as seen in Figure 7.
|
The user uploads their document set and ontology file by clicking the Upload button, activating the Upload subview, as seen in Figure 8. After confirming a selection, the upload process begins.
|
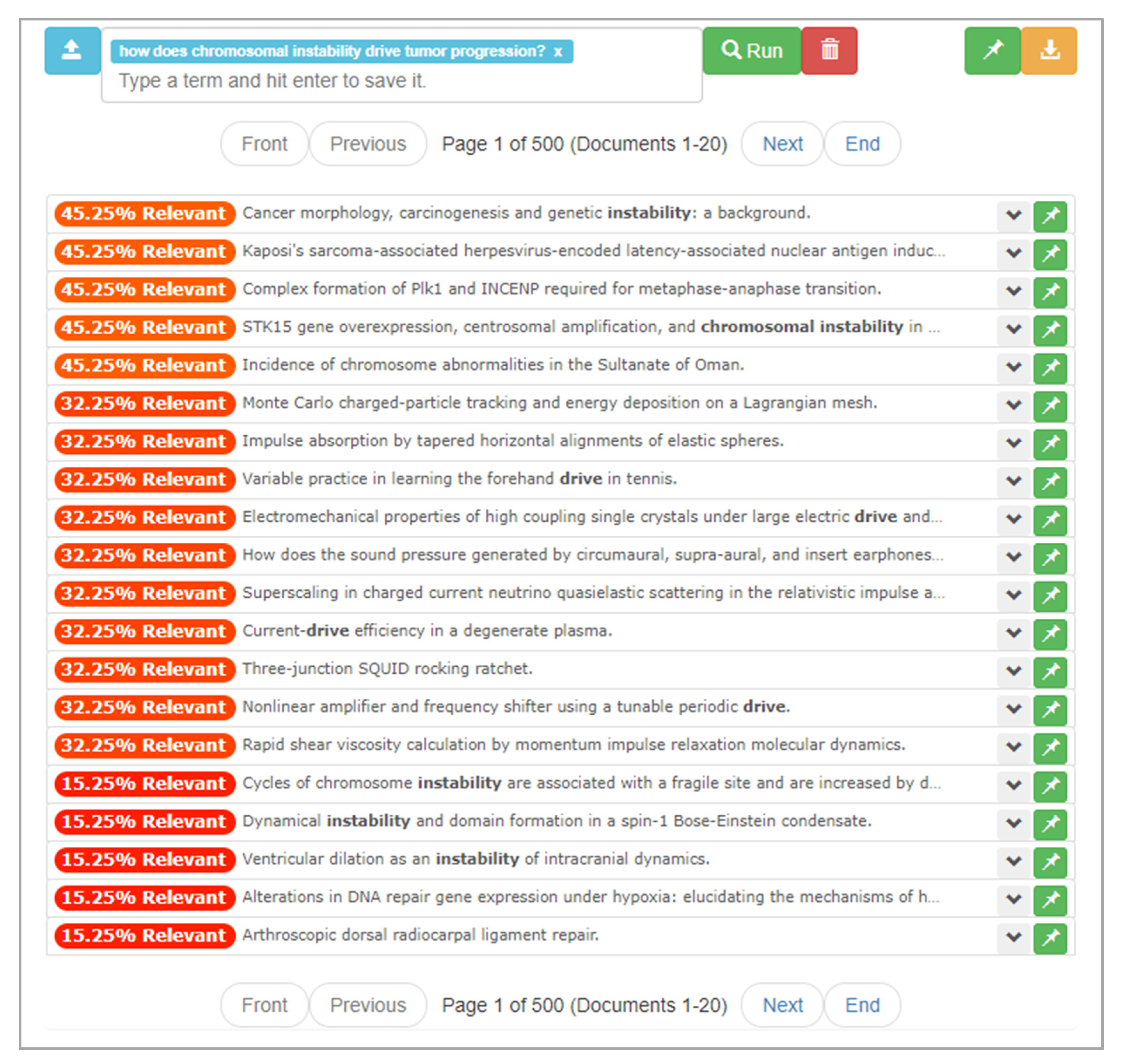
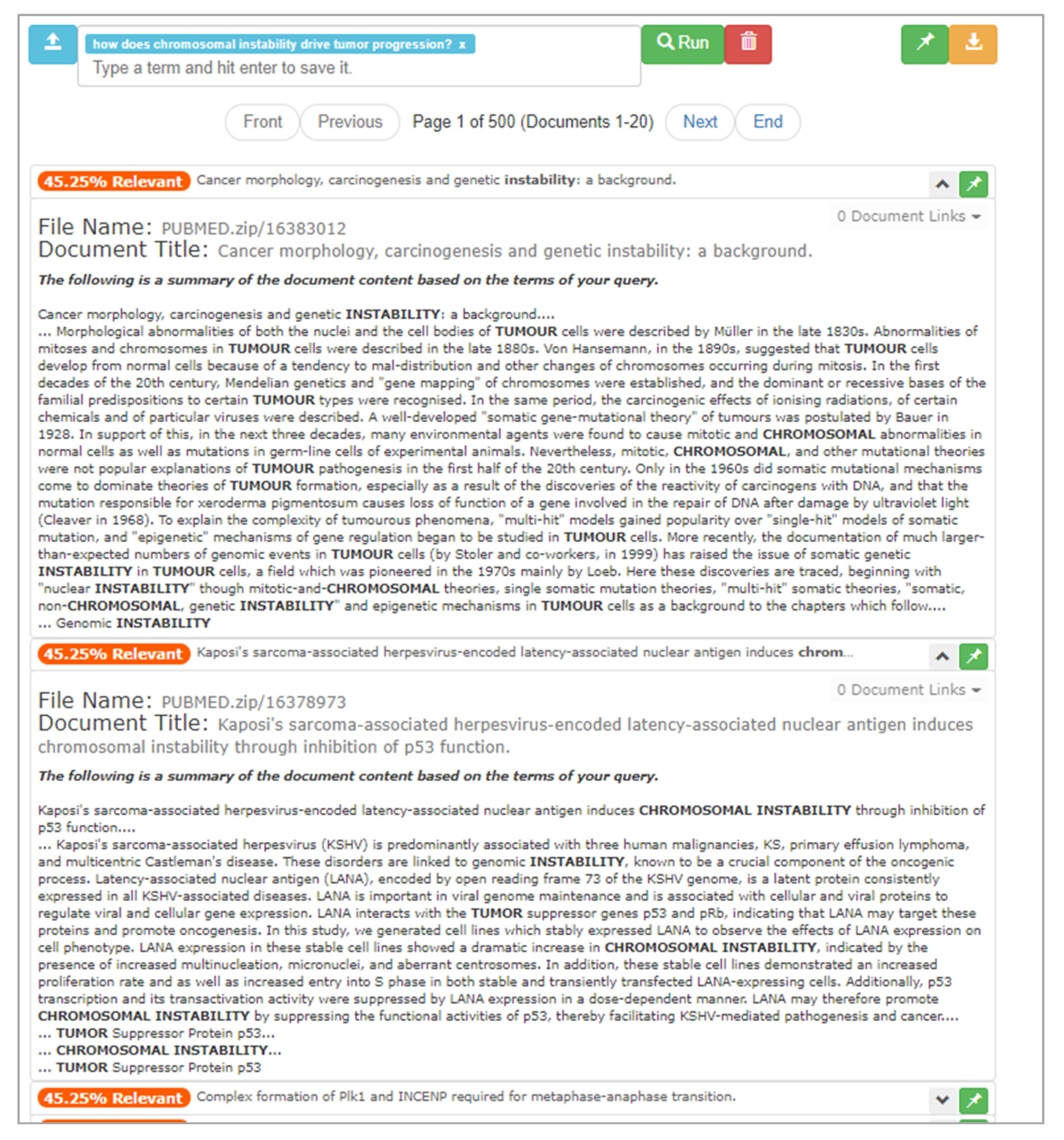
After the upload process is complete, the user begins typing the research question “How does chromosomal instability drive tumor progression?” into the textbox, finishing with a click of the “Run” button. In response, ONTSI provides the results of its computation, as seen in Figure 9. It presents the first page of 500 pages of documents, which totals 20 document entries. There are percentages to the left of each entry that use text and color to annotate the relevance of each document. This scalar is based on the cluster weightings within the dimensional space of the document set, where a 100% would be produced by documents within a cluster that aligns with every input feature. The scalar maintains a color scale between red and green, where red is at the zero point and green at 100%. For instance, at the top of the first page there are five documents that present an orange 45.25% relevance rating. Looking at these documents, the user scans the titles of the documents, where some have terms within their titles that relate to the research question.
|
Some documents at the top of the results could align with the user’s research question. To explore further, the user selects a few of the top documents, generating additional document information for inspection, as seen in Figure 10. Doing so, the user encounters a summarized version of their selected documents, which provides metadata and abstracts annotated with words and phrases related to the research question.
|
The user estimates that these top documents may align with their research question. Therefore, they click on the green “pin” button found at the rightmost point of each document to save their reference for future retrieval from the document set. These references are accessible by clicking the green “pin” button found at the top right of ONTSI to open the Saved List subview.
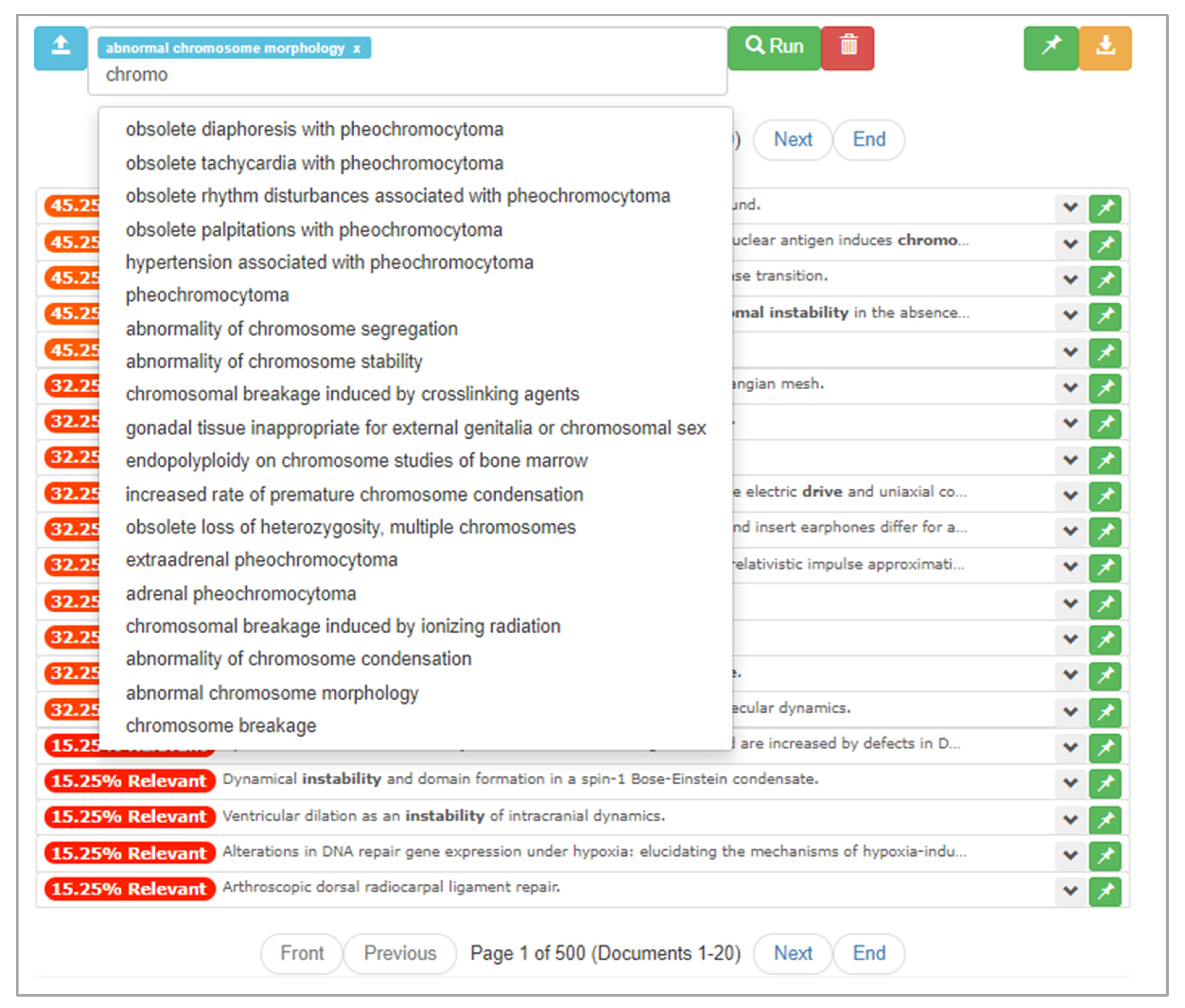
Although the user has now encountered some documents relevant to their research question, they choose to continue searching. This time, the user decides to take advantage of mediation opportunities when building their query. After closing the Saved List subview, the user begins a new search. After assessing the important words in their research question, the user types in the term “chromosomal.” At the point that they have typed “chromo,” they are presented with mediation opportunities, as seen in Figure 11. They inspect these mediation opportunities and add phenotypic terms that align with their research question.
|
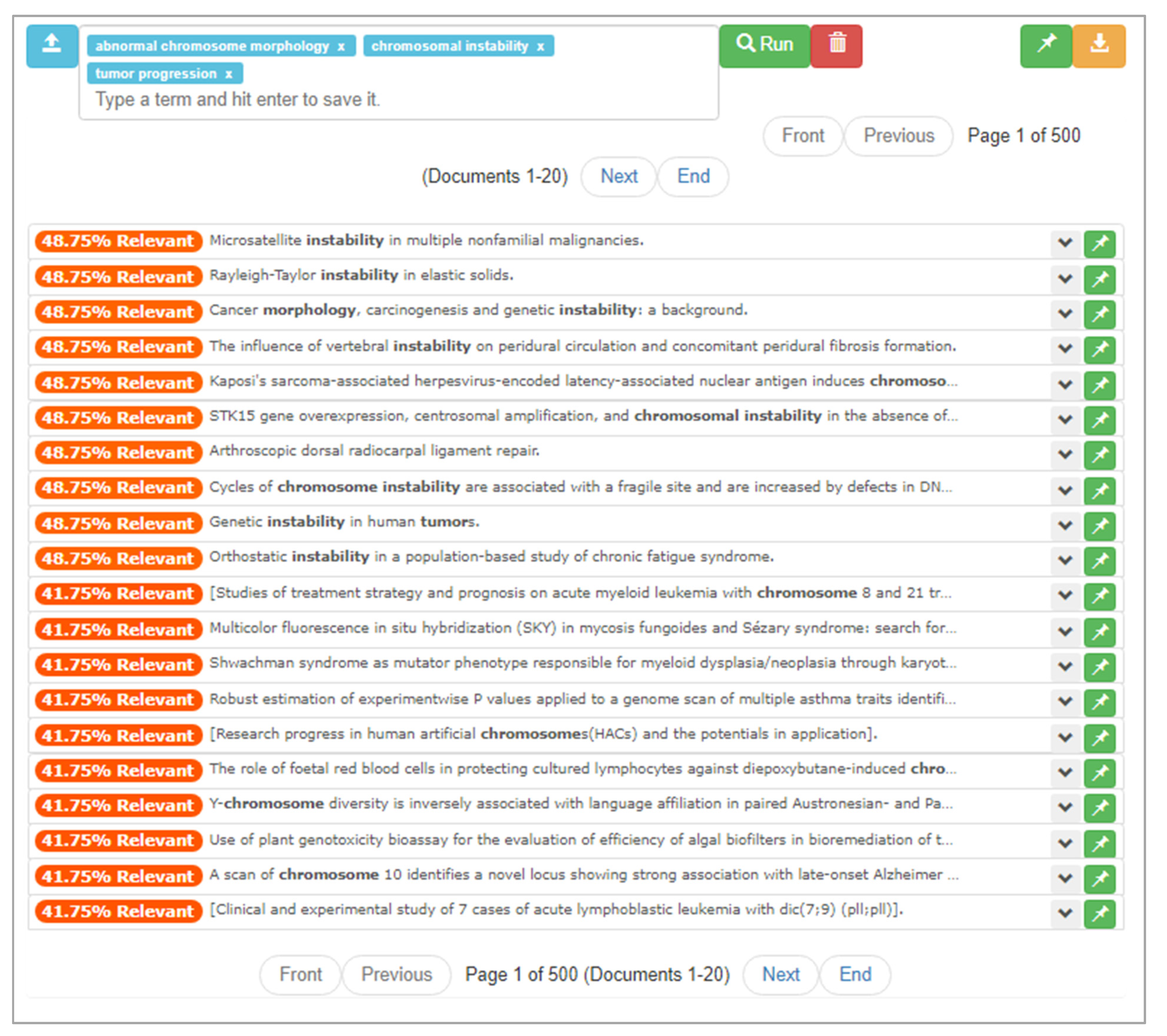
With the aid of mediation, the user builds a three-item query consisting of “abnormal chromosome morphology,” “chromosomal instability,” and “tumor progression.” After asking ONTSI to run with this query, the user encounters a set of results different from the one produced by their earlier search, as seen in Figure 12. Notably, an increased set of 10 documents at a 48.75% rating is encountered. Looking at these documents, the user notices some that are familiar, such as the saved “Genetic instability in human tumors.”
|
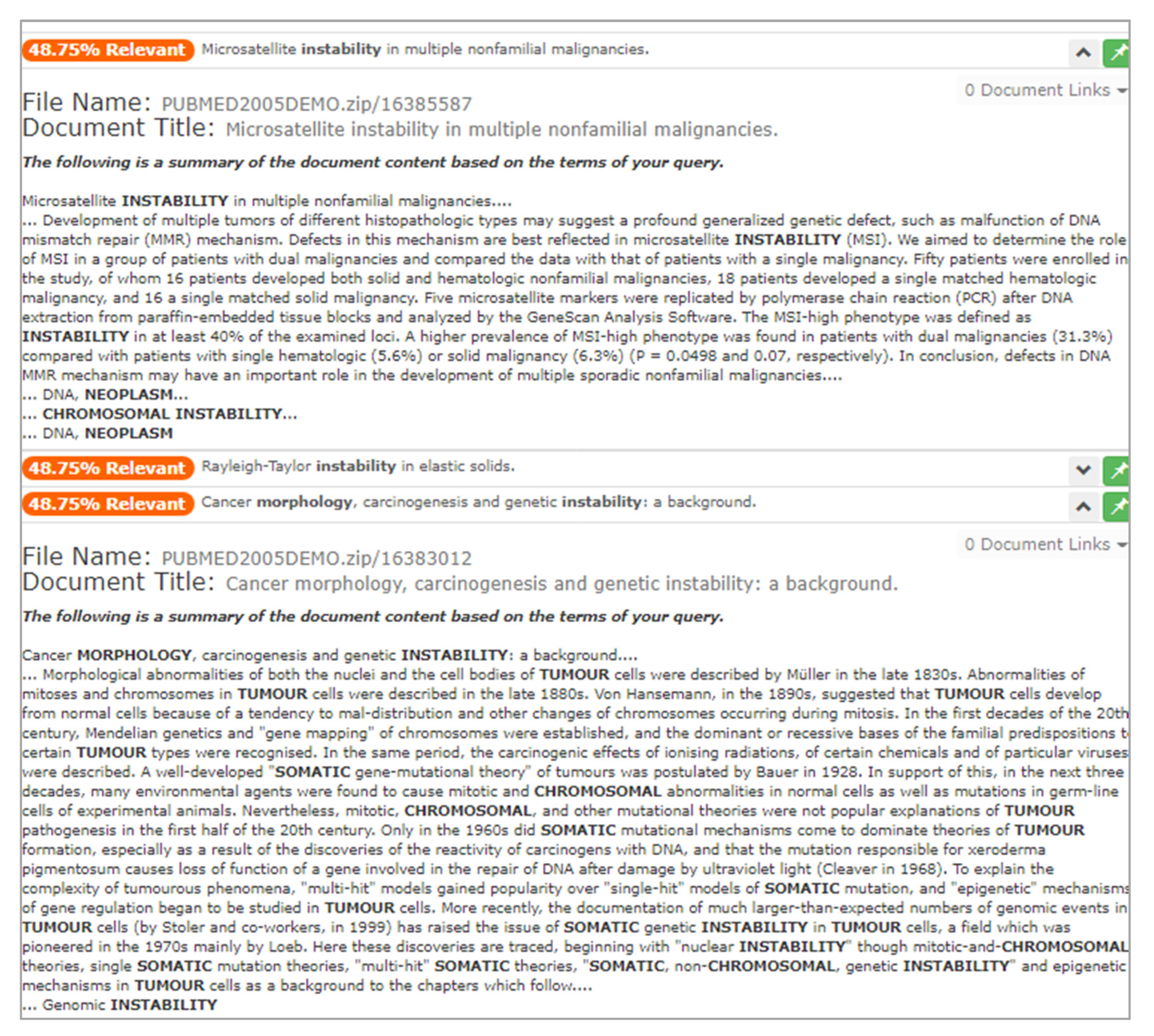
From this listing, the user selects two new documents for deeper inspection, as seen in Figure 13. They notice that terms such as “morphology” and “neoplasm” are now being highlighted within the document annotations. The adjusted query based on mediation opportunities has helped promote documents that align with their research question. In this case, the user finds value in the two documents, so they are saved.
|
Before concluding the search task, the user can upload a different ontology file to investigate how alternate vocabularies may bridge them to their document set. Their encounters could have also allowed them to make the assessment that the document set may not be best to help with their research question. If that is the case, they may upload a different document set, performing another scoping review. In any case, ONTSI provides a search task interface that has been generalized to support plug-and-play capabilities for user-provided ontology files and document sets, allowing users to customize the interface to match their search task objectives.
Discussion and conclusions
Evaluation of ONTSI
We have conducted ongoing, formative, task-driven user evaluations of ONTSI. These evaluations were informally conducted with a few people associated with our research lab; they have provided initial insights into how ontology-supported interfaces for health informatics can support users to perform elaborate search tasks involving large document sets. In these evaluations, we asked the users to perform a targeted set of tasks, such as researching questions outlined in the presented usage scenario. Initial sessions provided general insight into how users search and how ontologies can help mediate such tasks. From these sessions, we have learned a few things:
- Users are able to quickly transfer their experiences with previous interfaces to use ONTSI (e.g., A, B).
- Users are capable of utilizing ontology files to align their vocabulary with the vocabulary of the domain, even if they are not initially familiar with the ontology’s domain or its structure and content (e.g., C, D).
- Users are capable of understanding the requirements of the information-seeking process, expressing their valuation of the support they are provided by the interface as they performed their search tasks (e.g., E, F, G).
- Users felt that mediating ontologies make search tasks more manageable and easier, and not having them would negatively affect their task performance (e.g., E, F, G).
The following are some informal excerpts of some of the comments of those who have used ONTSI:
(A) “I think with ONTSI, I can immediately it matches my mental models of how I use search interfaces. I type things in, I click run, I go through pages of results.”
(B) “Once I understood what it was showing me, it helped me. Usually with new tools I tend to read through the documentation or watch videos. And then it still takes me like a while to pick up on them. Like, just running through them and using them a few times. Once you get the hang of it, usually you find success in whatever it’s providing you.”
(C) “But … you can get lost in the information too, right? So, if you have like so much so many things related in that ontology, it’s like, well, it can be useful. But it could also be a distraction for something that you know. There’s this flip side, but I think that’s on the searcher to know what they’re using and why they’re using it. So … for me to complete these tasks, if I hadn’t had the ontologies listed, then I would have had a much more difficult time. It essentially provided guidance … and a structure to something I was unfamiliar with in this case.”
(D) “I wasn’t necessarily intimidated, but I was just like—I don’t know what this is. But the background information for the context helped a little bit. A lot of big words, but they did help me when I was looking at the documents that I had to search for to find out which ones I felt best. So even though I did not have full understanding of the words, having them there in that background provided me a kind of help towards finding myself in the space of the question.”
(E) “I was thinking … where (the ontology) would have been helpful. So … it would have possibly brought up some of those other terms just from searching a few words and they would be able to make some connections between the text that was provided and some of my search terms (to see) … how relevant they were. So, if I was shooting in the dark and hoping for the best, which is what I was kind of doing (without the ontology), at the very least, it would have given you confidence of your actions. Yeah, I think so. A little bit more confidence.”
(F) “I thought it would be like pretty easy because I (am used to) answering … open questions like … find the things most relevant. So, this research question is for me … just an easier thing to do because I have background in doing that kind of stuff. ONTSI kind of functions like … a library tool that is available. This kind of tool felt very familiar to me. I wouldn’t say that I’m an expert when it comes to medical knowledge, but … I understand … basic terminology. So… what the terms meant or what they refer to wasn’t really … an issue. It wasn’t really alienating. I have like some general level of confidence just using the terms and trusting the tool as you went along.”
(G) “Yeah, this (ontology) would have helped because I can find the things that … share in common, and that can make it probably much easier to find the relevant documents. Yeah, being able to see the things that certain phrases … or words share in common. You can find that common link … that can find you the relevant documents.”
In the future, we plan to perform formal, empirical evaluations with users comparing ONTSI to other systems. Such evaluations will help generate new insights into features of interface designs and their qualitative and quantitative measures of how search task performances are affected. Beyond that, such evaluation studies may provide prescriptive guidelines for the design of optimal and effective interfaces for health informatics search tasks. In addition, we intend to further investigate how ontologies and machine learning should be integrated into elaborate and challenging search tasks that need domain-specific knowledge for optimal performance.
Limitations
The first limitation of ONTSI is the scaling of computational resources. ONTSI in its current state provides a plug-and-play experience that can handle the uploading and processing of both document sets and ontology files of large sizes. For instance, ONTSI easily handles HPO and its more than 11,000 ontology terms, alongside an extracted subset of PubMed of more than 10,000 documents. Yet, under the load of large-volume document sets and connected suites of ontology files, ONTSI’s computational systems may provide reduced responsiveness. To deal with such scenarios, further work is needed to solve overhead limitations—strategies such as pre-hosting common ontology files, establishing API connections to access externally hosted document sets, as well as simply expanding the computational power of our systems.
The second limitation of ONTSI is the support of ontology file formats. ONTSI in its current state can process the core encoded elements within the OWL format, a leading format for encoding ontologies. Yet, the format is quite verbose in its specification, requiring developments beyond the scope of our immediate research objectives. In addition, there are other formats used to encode ontologies that would be valuable to support ontology-supported interfaces for health informatics search tasks.
Conclusions
In summary, in this paper we began with an examination of the background on the topics of health informatics, machine learning, and ontologies. We then reviewed recent research on health informatics search tasks. Based on this review, we formalized a set of criteria for guiding designers when creating ontology-supported interfaces for health informatics search tasks involving large document sets. We then used these criteria to contrast traditional design strategies for interfaces of search tasks.
To demonstrate the utility of the criteria in the design process, we applied them to structure the creation of ONTSI (ONTology-supported Search Interface), an ontology-supported interface for health informatics search tasks involving large document sets. ONTSI combines five front-end subviews and two back-end computational systems. With these systems, ONTSI supplies a generalized interface that supports users’ ability to plug-and-play their provided document sets and an ontology file as a mediating resource within the interface when performing their health informatics search tasks.
The workflow of ONTSI was described and illustrated in a usage scenario. For our scenario, we used the Human Phenotype Ontology to mediate a search task on a subset of the PubMed document set. This usage scenario presented a narrative of a health professional performing a scoping review. Within the scenario, we found that ONTSI allows the user to utilize their ontology resource in a manner that aligns with both the unstructured and structured-like query expansion interface strategy. In the former, the user entered a research question without participating in mediation opportunities. In that case, ONTSI used HPO and WordNet as mediating resources to extend the user’s query within an expansion model to generate the results of a search task. In the latter case, the user took advantage of mediation opportunities during their query building. Although this usage scenario provides a single health informatics narrative, we believe value can be generated from both the criteria and ONTSI for health informatics in a broad sense. In this sense, we envision that our efforts can be further expanded to encompass tasks in informatics such as consumer informatics, nursing informatics, and ontology-supported domains beyond health and medicine, to name but a few.
In conclusion, in this paper we generated and proposed a set of criteria that can provide guidance to designers in creating ontology-supported interfaces for health informatics search tasks involving large document sets. We illustrated the utility of these criteria in the context of the creation and demonstration of ONTSI. We provided general insight from ongoing, formative, task-driven user evaluations of ONTSI. We hope to continue this research to promote the design of generalized ontology-supported interfaces for health informatics search tasks involving large document sets.
Acknowledgements
We would like to acknowledge NSERC and its support of our research.
Author contributions
Conceptualization, J.D. and K.S.; methodology, J.D. and K.S.; software, J.D.; validation, J.D. and K.S.; formal analysis, J.D. and K.S.; investigation, J.D. and K.S.; resources, Insight Lab, J.D. and K.S.; data curation, J.D.; writing—original draft preparation, J.D.; writing—review and editing, J.D. and K.S.; visualization, J.D.; supervision, K.S.; project administration, J.D.; funding acquisition, K.S. Both authors have read and agreed to the published version of the manuscript.
Funding
We acknowledge the support of the Natural Sciences and Engineering Research Council of Canada (NSERC).
Conflicts of interest
The authors declare no conflict of interest.
References
- ↑ 1.0 1.1 1.2 Wickramasinghe, Nilmini (2019/08). "Essential Considerations for Successful Consumer Health Informatics Solutions" (in en). Yearbook of Medical Informatics 28 (01): 158–164. doi:10.1055/s-0039-1677909. ISSN 0943-4747. PMC PMC6697544. PMID 31419828. http://www.thieme-connect.de/DOI/DOI?10.1055/s-0039-1677909.
- ↑ 2.0 2.1 2.2 Canadian Medical Association (2018). "The Future of Technology in Health and Health Care: A Primer". Canadian Medical Association. Archived from the original on 30 April 2019. https://web.archive.org/web/20190430220959/https://www.cma.ca/sites/default/files/pdf/health-advocacy/activity/2018-08-15-future-technology-health-care-e.pdf.
- ↑ 3.0 3.1 3.2 Demiris, G. (2016). "Consumer Health Informatics: Past, Present, and Future of a Rapidly Evolving Domain" (in en). Yearbook of Medical Informatics 25 (S 01): S42–S47. doi:10.15265/IYS-2016-s005. ISSN 0943-4747. PMC PMC5171509. PMID 27199196. http://www.thieme-connect.de/DOI/DOI?10.15265/IYS-2016-s005.
- ↑ 4.0 4.1 Zuccon, G.; Koopman, B. (2014). Goeuriot, L.; Jones, G.J.F.; Kelly, L. et al.. ed. "Integrating understandability in the evaluation of consumer health search engines". CEUR Workshop Proceedings 1276: 32–35. http://ceur-ws.org/Vol-1276/.
- ↑ 5.0 5.1 Chinese Journal of Cancer (1 December 2017). "The 150 most important questions in cancer research and clinical oncology series: questions 67–75: Edited by Chinese Journal of Cancer" (in en). Chinese Journal of Cancer 36 (1): 86, s40880–017–0254-z. doi:10.1186/s40880-017-0254-z. ISSN 1944-446X. PMC PMC5664810. PMID 29092716. https://cancercommun.biomedcentral.com/articles/10.1186/s40880-017-0254-z.
- ↑ Mehta, N.; Pandit, A. (1 June 2018). "Concurrence of big data analytics and healthcare: A systematic review" (in en). International Journal of Medical Informatics 114: 57–65. doi:10.1016/j.ijmedinf.2018.03.013. ISSN 1386-5056. https://www.sciencedirect.com/science/article/abs/pii/S1386505618302466.
- ↑ Thiébaut, Rodolphe; Cossin, Sébastien; Informatics, Section Editors for the IMIA Yearbook Section on Public Health and Epidemiology (2019/08). "Artificial Intelligence for Surveillance in Public Health" (in en). Yearbook of Medical Informatics 28 (01): 232–234. doi:10.1055/s-0039-1677939. ISSN 0943-4747. PMC PMC6697516. PMID 31419837. http://www.thieme-connect.de/DOI/DOI?10.1055/s-0039-1677939.
- ↑ 8.0 8.1 Saleemi, M.M.; Rodríguez, N.D.; Lilius, J.; Porres, I. (2011). "A Framework for Context-aware Applications for Smart Spaces". In Balandin, Sergey; Koucheryavy, Yevgeni; Hu, Honglin et al.. Smart Spaces and Next Generation Wired/Wireless Networking. Lecture notes in computer science. Heidelberg: Springer. pp. 14–25. ISBN 978-3-642-22874-2. OCLC 844916767. https://www.worldcat.org/title/mediawiki/oclc/844916767.
- ↑ 9.0 9.1 Gibson, C. J.; Dixon, B. E.; Abrams, K. (2015). "Convergent evolution of health information management and health informatics" (in en). Applied Clinical Informatics 06 (01): 163–184. doi:10.4338/ACI-2014-09-RA-0077. ISSN 1869-0327. PMC PMC4377568. PMID 25848421. http://www.thieme-connect.de/DOI/DOI?10.4338/ACI-2014-09-RA-0077.
- ↑ 10.0 10.1 10.2 Fang, Ruogu; Pouyanfar, Samira; Yang, Yimin; Chen, Shu-Ching; Iyengar, S. S. (14 June 2016). "Computational Health Informatics in the Big Data Age: A Survey". ACM Computing Surveys 49 (1): 12:1–12:36. doi:10.1145/2932707. ISSN 0360-0300. https://doi.org/10.1145/2932707.
- ↑ 11.0 11.1 11.2 Köhler, Sebastian; Carmody, Leigh; Vasilevsky, Nicole; Jacobsen, Julius O B; Danis, Daniel; Gourdine, Jean-Philippe; Gargano, Michael; Harris, Nomi L et al. (8 January 2019). "Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources". Nucleic Acids Research 47 (D1): D1018–D1027. doi:10.1093/nar/gky1105. ISSN 0305-1048. PMC PMC6324074. PMID 30476213. https://doi.org/10.1093/nar/gky1105.
- ↑ 12.0 12.1 12.2 Carayon, Pascale; Hoonakker, Peter (2019/08). "Human Factors and Usability for Health Information Technology: Old and New Challenges" (in en). Yearbook of Medical Informatics 28 (01): 071–077. doi:10.1055/s-0039-1677907. ISSN 0943-4747. PMC PMC6697515. PMID 31419818. http://www.thieme-connect.de/DOI/DOI?10.1055/s-0039-1677907.
- ↑ Gamache, Roland; Kharrazi, Hadi; Weiner, Jonathan P. (2018/08). "Public and Population Health Informatics: The Bridging of Big Data to Benefit Communities" (in en). Yearbook of Medical Informatics 27 (01): 199–206. doi:10.1055/s-0038-1667081. ISSN 0943-4747. PMC PMC6115205. PMID 30157524. http://www.thieme-connect.de/DOI/DOI?10.1055/s-0038-1667081.
- ↑ Brewer, LaPrincess C.; Fortuna, Karen L.; Jones, Clarence; Walker, Robert; Hayes, Sharonne N.; Patten, Christi A.; Cooper, Lisa A. (14 January 2020). "Back to the Future: Achieving Health Equity Through Health Informatics and Digital Health" (in EN). JMIR mHealth and uHealth 8 (1): e14512. doi:10.2196/14512. PMC PMC6996775. PMID 31934874. https://mhealth.jmir.org/2020/1/e14512.
- ↑ 15.0 15.1 Wu, Charley M.; Meder, Björn; Filimon, Flavia; Nelson, Jonathan D. (1 August 2017). "Asking better questions: How presentation formats influence information search." (in en). Journal of Experimental Psychology: Learning, Memory, and Cognition 43 (8): 1274–1297. doi:10.1037/xlm0000374. ISSN 1939-1285. http://doi.apa.org/getdoi.cfm?doi=10.1037/xlm0000374.
- ↑ 16.0 16.1 Talbot, Justin; Lee, Bongshin; Kapoor, Ashish; Tan, Desney S. (4 April 2009). "EnsembleMatrix: Interactive visualization to support machine learning with multiple classifiers" (in en). Proceedings of the SIGCHI Conference on Human Factors in Computing Systems (Boston MA USA: ACM): 1283–1292. doi:10.1145/1518701.1518895. ISBN 978-1-60558-246-7. https://dl.acm.org/doi/10.1145/1518701.1518895.
- ↑ 17.0 17.1 Hohman, Fred; Kahng, Minsuk; Pienta, Robert; Chau, Duen Horng (1 August 2019). "Visual Analytics in Deep Learning: An Interrogative Survey for the Next Frontiers". IEEE Transactions on Visualization and Computer Graphics 25 (8): 2674–2693. doi:10.1109/TVCG.2018.2843369. ISSN 1941-0506. PMC PMC6703958. PMID 29993551. https://ieeexplore.ieee.org/document/8371286/.
- ↑ 18.0 18.1 Yuan, Jun; Chen, Changjian; Yang, Weikai; Liu, Mengchen; Xia, Jiazhi; Liu, Shixia (25 November 2020). "A survey of visual analytics techniques for machine learning" (in en). Computational Visual Media 7 (1): 3–36. doi:10.1007/s41095-020-0191-7. ISSN 2096-0433. https://doi.org/10.1007/s41095-020-0191-7.
- ↑ 19.0 19.1 Endert, A.; Ribarsky, W.; Turkay, C.; Wong, B. L. William; Nabney, I.; Blanco, I. Díaz; Rossi, F. (2017). "The State of the Art in Integrating Machine Learning into Visual Analytics" (in en). Computer Graphics Forum 36 (8): 458–486. doi:10.1111/cgf.13092. ISSN 1467-8659. https://onlinelibrary.wiley.com/doi/abs/10.1111/cgf.13092.
- ↑ 20.0 20.1 Jusoh, S; Awajan, A; Obeid, N (1 May 2020). "The Use of Ontology in Clinical Information Extraction". Journal of Physics: Conference Series 1529: 052083. doi:10.1088/1742-6596/1529/5/052083. ISSN 1742-6588. https://iopscience.iop.org/article/10.1088/1742-6596/1529/5/052083.
- ↑ 21.0 21.1 Lytvyn, Vasyl; Dosyn, Dmytro; Vysotska, Victoria; Hryhorovych, Andrii (1 August 2020). "Method of Ontology Use in OODA". 2020 IEEE Third International Conference on Data Stream Mining & Processing (DSMP) (Lviv, Ukraine: IEEE): 409–413. doi:10.1109/DSMP47368.2020.9204107. ISBN 978-1-7281-3214-3. https://ieeexplore.ieee.org/document/9204107/.
- ↑ 22.0 22.1 Román-Villarán, E.; Pérez-Leon, F. P.; Escobar-Rodriguez, G. A.; Martínez-García, A.; Álvarez-Romero, C.; Parra-Calderón, C. L. (21 August 2019). "An Ontology-Based Personalized Decision Support System for Use in the Complex Chronically Ill Patient". Studies in Health Technology and Informatics 264: 758–762. doi:10.3233/SHTI190325. ISSN 1879-8365. PMID 31438026. https://pubmed.ncbi.nlm.nih.gov/31438026.
- ↑ Sacha, D.; Sedlmair, M.; Zhang L.et al. (2016). "Human-centered Machine Learning Through Interactive Visualization: Review and Open Challenges". In Verleysen, Michel. 24th European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning: ESANN 2016. ESSAN, Université catholique de Louvain, Katholieke Universiteit Leuven. Louvain-la-Neuve, Belgique: Ciaco - i6doc.com. pp. 641–646. ISBN 978-2-87587-027-8. OCLC 964654436. https://www.worldcat.org/title/mediawiki/oclc/964654436.
- ↑ Amershi, Saleema; Cakmak, Maya; Knox, William Bradley; Kulesza, Todd (22 December 2014). "Power to the People: The Role of Humans in Interactive Machine Learning" (in en). AI Magazine 35 (4): 105–120. doi:10.1609/aimag.v35i4.2513. ISSN 2371-9621. https://ojs.aaai.org/index.php/aimagazine/article/view/2513.
- ↑ 25.0 25.1 25.2 Zeng, Qing T.; Tse, Tony (1 January 2006). "Exploring and Developing Consumer Health Vocabularies". Journal of the American Medical Informatics Association 13 (1): 24–29. doi:10.1197/jamia.M1761. ISSN 1067-5027. PMC PMC1380193. PMID 16221948. https://doi.org/10.1197/jamia.M1761.
- ↑ 26.0 26.1 26.2 26.3 Arp, Robert; Smith, Barry; Spear, Andrew D. (2015). Building ontologies with Basic Formal Ontology. Cambridge, Massachusetts: Massachusetts Institute of Technology. ISBN 978-0-262-52781-1.
- ↑ Bikakis, N.; Sellis, T. (2016). "Exploration and Visualization in the Web of Big Linked Data: A Survey of the State of the Art". arXiv. arXiv:1601.08059. https://arxiv.org/abs/1601.08059.
- ↑ Carpendale, Sheelagh; Chen, Min; Evanko, Daniel; Gehlenborg, Nils; Görg, Carsten; Hunter, Larry; Rowland, Francis; Storey, Margaret-Anne et al. (1 March 2014). "Ontologies in Biological Data Visualization". IEEE Computer Graphics and Applications 34 (2): 8–15. doi:10.1109/MCG.2014.33. ISSN 1558-1756. https://ieeexplore.ieee.org/document/6777435/.
- ↑ Dou, Dejing; Wang, Hao; Liu, Haishan (1 February 2015). "Semantic data mining: A survey of ontology-based approaches". Proceedings of the 2015 IEEE 9th International Conference on Semantic Computing (IEEE ICSC 2015) (Anaheim, CA, USA: IEEE): 244–251. doi:10.1109/ICOSC.2015.7050814. ISBN 978-1-4799-7935-6. http://ieeexplore.ieee.org/document/7050814/.
- ↑ Livingston, Kevin M.; Bada, Michael; Baumgartner, William A.; Hunter, Lawrence E. (23 April 2015). "KaBOB: ontology-based semantic integration of biomedical databases". BMC Bioinformatics 16 (1): 126. doi:10.1186/s12859-015-0559-3. ISSN 1471-2105. PMC PMC4448321. PMID 25903923. https://doi.org/10.1186/s12859-015-0559-3.
- ↑ Salkic, S.; Softic, S.; Taraghi, B. Ebner, M. (2015). "Linked data driven visual analytics for tracking learners in a PLE". In Pongratz, Hans; Gesellschaft für Informatik. DeLFI 2015 - die 13. E-Learning Fachtagung Informatik der Gesellschaft für Informatik e.V: 1.-4. September 2015 München, Deutschland. GI-Edition Lecture Notes in Informatics Proceedings. Bonn: Ges. für Informatik. pp. 329–331. ISBN 978-3-88579-641-1. OCLC 927803453. https://www.worldcat.org/title/mediawiki/oclc/927803453.
- ↑ Jakus, Grega; Milutinović, Veljko; Omerović, Sanida; Tomažič, Sašo (2013). Concepts, ontologies, and knowledge representation. SpringerBriefs in computer science. New York: Springer. ISBN 978-1-4614-7821-8. OCLC 841495258. https://www.worldcat.org/title/mediawiki/oclc/841495258.
- ↑ Rector, A.; Schulz, S.; Rodrigues, J.M. et al. (1 January 2019). "On beyond Gruber: “Ontologies” in today’s biomedical information systems and the limits of OWL" (in en). Journal of Biomedical Informatics 100 Suppl.. doi:10.1016/j.yjbinx.2019.100002. ISSN 1532-0464. https://www.sciencedirect.com/science/article/pii/S2590177X19300010.
- ↑ Rodríguez, Natalia Díaz; Cuéllar, M. P.; Lilius, Johan; Calvo-Flores, Miguel Delgado (1 April 2014). "A survey on ontologies for human behavior recognition" (in en). ACM Computing Surveys 46 (4): 1–33. doi:10.1145/2523819. ISSN 0360-0300. https://dl.acm.org/doi/10.1145/2523819.
- ↑ Katifori, Akrivi; Torou, Elena; Vassilakis, Costas; Lepouras, Georgios; Halatsis, Constantin (1 June 2008). "Selected results of a comparative study of four ontology visualization methods for information retrieval tasks". 2008 Second International Conference on Research Challenges in Information Science (Marrakech: IEEE): 133–140. doi:10.1109/RCIS.2008.4632101. ISBN 978-1-4244-1677-6. http://ieeexplore.ieee.org/document/4632101/.
- ↑ Raghupathi, Wullianallur; Raghupathi, Viju (7 February 2014). "Big data analytics in healthcare: promise and potential" (in en). Health Information Science and Systems 2 (1). doi:10.1186/2047-2501-2-3. ISSN 2047-2501. PMC PMC4341817. PMID 25825667. https://doi.org/10.1186/2047-2501-2-3.
- ↑ 37.0 37.1 Russell-Rose, T.; Chamberlain, J.; Azzopardi, L. (2018). "Information retrieval in the workplace: A comparison of professional search practices" (in en). Information Processing & Management 54 (6): 1042–1057. doi:10.1016/j.ipm.2018.07.003. ISSN 0306-4573. https://www.sciencedirect.com/science/article/abs/pii/S0306457318300220.
- ↑ Russell-Rose, Tony; Chamberlain, Jon (2 October 2017). "Expert Search Strategies: The Information Retrieval Practices of Healthcare Information Professionals" (in EN). JMIR Medical Informatics 5 (4): e7680. doi:10.2196/medinform.7680. PMC PMC5643841. PMID 28970190. https://medinform.jmir.org/2017/4/e33.
- ↑ 39.0 39.1 Huurdeman, H.C. (2017). "Dynamic Compositions: Recombining Search User Interface Features for Supporting Complex Work Tasks". CEUR Workshop Proceedings 1798: 22–25. http://ceur-ws.org/Vol-1798/.
- ↑ Zahabi, Maryam; Kaber, David B.; Swangnetr, Manida (1 August 2015). "Usability and Safety in Electronic Medical Records Interface Design: A Review of Recent Literature and Guideline Formulation" (in en). Human Factors 57 (5): 805–834. doi:10.1177/0018720815576827. ISSN 0018-7208. https://doi.org/10.1177/0018720815576827.
- ↑ Dudley, John J.; Kristensson, Per Ola (13 June 2018). "A Review of User Interface Design for Interactive Machine Learning". ACM Transactions on Interactive Intelligent Systems 8 (2): 8:1–8:37. doi:10.1145/3185517. ISSN 2160-6455. https://doi.org/10.1145/3185517.
- ↑ 42.0 42.1 Harvey, Morgan; Hauff, Claudia; Elsweiler, David (9 August 2015). "Learning by Example: Training Users with High-quality Query Suggestions" (in en). Proceedings of the 38th International ACM SIGIR Conference on Research and Development in Information Retrieval (Santiago Chile: ACM): 133–142. doi:10.1145/2766462.2767731. ISBN 978-1-4503-3621-5. https://dl.acm.org/doi/10.1145/2766462.2767731.
- ↑ Soldaini, Luca; Yates, Andrew; Yom-Tov, Elad; Frieder, Ophir; Goharian, Nazli (16 July 2015). "Enhancing web search in the medical domain via query clarification" (in en). Information Retrieval Journal 19 (1-2): 149–173. doi:10.1007/s10791-015-9258-y. ISSN 1386-4564. https://doi.org/10.1007/s10791-015-9258-y.
- ↑ Anderson, James D; Wischgoll, Thomas (26 January 2020). "Visualization of Search Results of Large Document Sets". Electronic Imaging 2020 (1): 388–1–388-7. doi:10.2352/ISSN.2470-1173.2020.1.VDA-388. https://www.ingentaconnect.com/content/ist/ei/2020/00002020/00000001/art00006;jsessionid=1ui77rulkmwwg.x-ic-live-03.
- ↑ Soldaini, Luca; Cohan, Arman; Yates, Andrew; Goharian, Nazli; Frieder, Ophir (2015), Hanbury, Allan; Kazai, Gabriella; Rauber, Andreas et al.., eds., "Retrieving Medical Literature for Clinical Decision Support" (in en), Advances in Information Retrieval (Cham: Springer International Publishing) 9022: 538–549, doi:10.1007/978-3-319-16354-3_59, ISBN 978-3-319-16353-6, http://link.springer.com/10.1007/978-3-319-16354-3_59. Retrieved 2021-09-23
- ↑ Wilson, Max L. (2012). Search user interface design. Synthesis lectures on information concepts, retrieval, and services. San Rafael, Calif.: Morgan & Claypool. ISBN 978-1-60845-689-5. OCLC 780340844. https://www.worldcat.org/title/mediawiki/oclc/780340844.
- ↑ Zielstorff, R.D. (2003). "Controlled vocabularies for consumer health" (in en). Journal of Biomedical Informatics 36 (4-5): 326–333. doi:10.1016/j.jbi.2003.09.015. ISSN 1532-0464. https://www.sciencedirect.com/science/article/pii/S1532046403000960.
- ↑ 48.0 48.1 National Center for Biotechnology Information. "PubMed.gov". National Institutes of Health. https://pubmed.ncbi.nlm.nih.gov/. Retrieved 18 January 2021.
- ↑ McCray, Alexa T.; Tse, Tony (2003). "Understanding Search Failures in Consumer Health Information Systems". AMIA Annual Symposium Proceedings 2003: 430–434. ISSN 1942-597X. PMC 1479930. PMID 14728209. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1479930/.
- ↑ Keselman, Alla; Browne, Allen C.; Kaufman, David R. (1 July 2008). "Consumer Health Information Seeking as Hypothesis Testing". Journal of the American Medical Informatics Association 15 (4): 484–495. doi:10.1197/jamia.M2449. ISSN 1067-5027. PMC PMC2442260. PMID 18436912. https://doi.org/10.1197/jamia.M2449.
- ↑ Luo, Jake; Wu, Min; Gopukumar, Deepika; Zhao, Yiqing (1 January 2016). "Big Data Application in Biomedical Research and Health Care: A Literature Review" (in en). Biomedical Informatics Insights 8: BII.S31559. doi:10.4137/BII.S31559. ISSN 1178-2226. PMC PMC4720168. PMID 26843812. https://doi.org/10.4137/BII.S31559.
- ↑ Qvarfordt, Pernilla; Golovchinsky, Gene; Dunnigan, Tony; Agapie, Elena (28 July 2013). "Looking ahead: query preview in exploratory search" (in en). Proceedings of the 36th international ACM SIGIR conference on Research and development in information retrieval (Dublin Ireland: ACM): 243–252. doi:10.1145/2484028.2484084. ISBN 978-1-4503-2034-4. https://dl.acm.org/doi/10.1145/2484028.2484084.
- ↑ 53.0 53.1 Jimmy; Zuccon, Guido; Koopman, Bevan (2018), Pasi, Gabriella; Piwowarski, Benjamin; Azzopardi, Leif et al.., eds., "Choices in Knowledge-Base Retrieval for Consumer Health Search", Advances in Information Retrieval (Cham: Springer International Publishing) 10772: 72–85, doi:10.1007/978-3-319-76941-7_6, ISBN 978-3-319-76940-0, http://link.springer.com/10.1007/978-3-319-76941-7_6. Retrieved 2021-09-23
- ↑ 54.0 54.1 Azad, H.K.; Deepak, A. (2019). "A new approach for query expansion using Wikipedia and WordNet" (in en). Information Sciences 492: 147–163. doi:10.1016/j.ins.2019.04.019. ISSN 0020-0255. https://www.sciencedirect.com/science/article/abs/pii/S0020025519303263.
- ↑ Jimmy, J.; Zuccon, G.; Palotti, J. et al. (2018). "Overview of the CLEF 2018 Consumer Health Search Task". CEUR Workshop Proceedings 2125: 1–15. http://ceur-ws.org/Vol-2125/.
- ↑ Capuano, Nicola; Longhi, Andrea; Salerno, Saverio; Toti, Daniele (4 May 2015). "Ontology-driven Generation of Training Paths in the Legal Domain" (in en). International Journal of Emerging Technologies in Learning (iJET) 10 (7): 14–22. doi:10.3991/ijet.v10i7.4609. ISSN 1863-0383. https://online-journals.org/index.php/i-jet/article/view/4609.
- ↑ 57.0 57.1 Köhler, Sebastian; Gargano, Michael; Matentzoglu, Nicolas; Carmody, Leigh C; Lewis-Smith, David; Vasilevsky, Nicole A; Danis, Daniel; Balagura, Ganna et al. (8 January 2021). "The Human Phenotype Ontology in 2021" (in en). Nucleic Acids Research 49 (D1): D1207–D1217. doi:10.1093/nar/gkaa1043. ISSN 0305-1048. PMC PMC7778952. PMID 33264411. https://academic.oup.com/nar/article/49/D1/D1207/6017351.
- ↑ Lüke, Thomas; Schaer, Philipp; Mayr, Philipp (2012), Zaphiris, Panayiotis; Buchanan, George; Rasmussen, Edie et al.., eds., "Improving Retrieval Results with Discipline-Specific Query Expansion", Theory and Practice of Digital Libraries (Berlin, Heidelberg: Springer Berlin Heidelberg) 7489: 408–413, doi:10.1007/978-3-642-33290-6_44, ISBN 978-3-642-33289-0, http://link.springer.com/10.1007/978-3-642-33290-6_44. Retrieved 2021-09-23
- ↑ Jay, Caroline; Harper, Simon; Dunlop, Ian; Smith, Sam; Sufi, Shoaib; Goble, Carole; Buchan, Iain (14 January 2016). "Natural Language Search Interfaces: Health Data Needs Single-Field Variable Search" (in EN). Journal of Medical Internet Research 18 (1): e4912. doi:10.2196/jmir.4912. PMC PMC4731680. PMID 26769334. https://www.jmir.org/2016/1/e13.
- ↑ 60.0 60.1 60.2 "Apache Solr". Apache Software Foundation. https://solr.apache.org/. Retrieved 18 January 2021.
- ↑ Bostock, M.. "D3.js". https://d3js.org/. Retrieved 18 January 2021.
- ↑ Köhler, Sebastian; Doelken, Sandra C.; Mungall, Christopher J.; Bauer, Sebastian; Firth, Helen V.; Bailleul-Forestier, Isabelle; Black, Graeme C. M.; Brown, Danielle L. et al. (1 January 2014). "The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data". Nucleic Acids Research 42 (D1): D966–D974. doi:10.1093/nar/gkt1026. ISSN 0305-1048. PMC PMC3965098. PMID 24217912. https://doi.org/10.1093/nar/gkt1026.
- ↑ Köhler, S.; Robinson, P.. "HPO Web Browser: Blindness Infopage". Human Phenotype Ontology Project. http://compbio.charite.de/hpoweb/showterm?id=HP:0000618. Retrieved 18 January 2021.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. Some grammar and punctuation was cleaned up to improve readability. In some cases important information was missing from the references, and that information was added. The original article uses the web address to cite the Human Phenotype Ontology; for this version the HPO-recommended citation of the 2021 article in Nucleic Acids Research was used.