Difference between revisions of "Journal:Development of Biosearch System for biobank management and storage of disease-associated genetic information"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 23: | Line 23: | ||
'''Objective''': Databases and software are important to manage modern high-throughput [[Laboratory|laboratories]] and store clinical and [[Genome informatics|genomic information]] for [[quality assurance]]. Commercial software is expensive, with proprietary code issues, while academic versions have adaptation issues. Our aim was to develop an adaptable in-house software system that can store specimen- and disease-associated genetic information in [[biobank]]s to facilitate [[translational research]]. | '''Objective''': Databases and software are important to manage modern high-throughput [[Laboratory|laboratories]] and store clinical and [[Genome informatics|genomic information]] for [[quality assurance]]. Commercial software is expensive, with proprietary code issues, while academic versions have adaptation issues. Our aim was to develop an adaptable in-house software system that can store specimen- and disease-associated genetic information in [[biobank]]s to facilitate [[translational research]]. | ||
'''Methods''': A prototype was designed per the research requirements, and computational tools were used to develop the software under three tiers, using Visual Basic and ASP.net for the presentation tier, SQL Server for the data tier, and Ajax and JavaScript for the business tier. We retrieved specimens from the biobank using this software and performed microarray-based transcriptomic analysis to detect | '''Methods''': A prototype was designed per the research requirements, and computational tools were used to develop the software under three tiers, using Visual Basic and ASP.net for the presentation tier, SQL Server for the data tier, and Ajax and JavaScript for the business tier. We retrieved specimens from the biobank using this software and performed microarray-based transcriptomic analysis to detect differentially expressed genes (DEGs) with FC ±2 and ''P''-value <0.05 in triple-negative breast [[cancer]] cases. The Ingenuity Pathway Analysis (IPA) tool was used to predict canonical molecular pathways associated with disease. Overall performance and utility of software was evaluated by Apache JMeter software, CRUD function testing, and a set of user feedback questionnaires. | ||
'''Results''': We developed Biosearch System, a web-based [[laboratory information management system]] (LIMS) enabling management of biobank samples (e.g., tissue, blood, FTTP slides) and their extracts (e.g., DNA, RNA, and proteins) with clinical and experimental details. The client satisfaction feedback was excellent, with a score of 4.7 out of 5. We identified a total of 1,181 DEGs, including both upregulated (IFI6, LEF1, FANCI, CASC5, PLXNA3, etc.) and down-regulated (ADH1B, LYVE1, ADH1C, ADH1B, ADIPOQ, PLIN1, LYVE1, etc.) genes in triple-negative breast cancer. Pathway analysis of DEGs revealed significant activation of interferon signaling (z-score 2.646) and kinetochore metaphase signaling pathway (z-score 2.138) in cancer. | '''Results''': We developed Biosearch System, a web-based [[laboratory information management system]] (LIMS) enabling management of biobank samples (e.g., tissue, blood, FTTP slides) and their extracts (e.g., DNA, RNA, and proteins) with clinical and experimental details. The client satisfaction feedback was excellent, with a score of 4.7 out of 5. We identified a total of 1,181 DEGs, including both upregulated (IFI6, LEF1, FANCI, CASC5, PLXNA3, etc.) and down-regulated (ADH1B, LYVE1, ADH1C, ADH1B, ADIPOQ, PLIN1, LYVE1, etc.) genes in triple-negative breast cancer. Pathway analysis of DEGs revealed significant activation of interferon signaling (z-score 2.646) and kinetochore metaphase signaling pathway (z-score 2.138) in cancer. | ||
| Line 68: | Line 68: | ||
===Database structure=== | ===Database structure=== | ||
A normal relational database is used for the system. Information is stored in specific tables like the <tt>SampleInfo</tt> table, which contains the sample's information, the <tt>ProductInfo</tt> table, which contains the type of sample information (e.g., DNA, RNA), the <tt>PatientInfo</tt> table, which contains a patient's related information, the <tt>SampleStorage</tt> table, which contains the storage location information (e.g., refrigerators, shelf, box container), and the <tt>UsersInfo</tt> table, which contains user-related information. The type of relation is one-to-many (one patient::multi-samples, one sample::multi-products, one refrigerator::multi-samples, etc.). | A normal relational database is used for the system. Information is stored in specific tables like the <tt>SampleInfo</tt> table, which contains the sample's information, the <tt>ProductInfo</tt> table, which contains the type of sample information (e.g., DNA, RNA), the <tt>PatientInfo</tt> table, which contains a patient's related information, the <tt>SampleStorage</tt> table, which contains the storage location information (e.g., refrigerators, shelf, box container), and the <tt>UsersInfo</tt> table, which contains user-related information. The type of relation is one-to-many (one patient::multi-samples, one sample::multi-products, one refrigerator::multi-samples, etc.). | ||
===Workflow and biobank organizational structure=== | |||
The software can only be used with an authorized username and password, and after successful login on a web-based client PC, users are allowed to add new data, update/edit data, follow up with patients, and retrieve data for analysis and interpretation. Biosearch System also manages the storage, quality, quantity, distribution, and maintenance of specimens (e.g., tissue, blood, serum, etc.) and its derivatives (e.g., DNA, RNA, protein, plasma, etc.). Per the existing CEGMR biobank organizational structure, assigned users have different level of rights for using the software. Researchers are end users of the biobank; they request samples based on their active research projects. Dedicated biobank staff examine the request and update the status, handing it off to the supervisor, who approves or rejects the request with valid reason. However, the final decision is made by the director, who has ultimate authority to reconsider the request or reverse the supervisors' decision. Once approved by the director, the biobank delivers the requested items within a week. Reminders for pending tasks are emailed to relevant staff until the final decision is made. This system enables researchers full access to the biobank information by allowing for the management of specimens and samples, the notification for request approval or rejection, and provision of the overall summary of the biobank’s inventory to researchers. | |||
===Technical procedure for linking clinical information to biobank specimens=== | |||
The LIMS manages all the relevant information for all deposited patients’ specimens and samples for systematic clinical research. We use a medical record number (MRN) as the primary key and a biobank number as the secondary key to connect two sections of the database. Patients’ samples are stored at the biobank with a unique allocated biobank number, which consists of 10 digits formatted as sample type (XX), serial number (XXXX), year (XX), and extraction type (XX). Sample types in our database includes AM (amniotic fluid), BL (blood), BO (bone marrow), CO (cord blood), CS (cervix swab), LN (lymph node), PC (product of conception), PL (paraffin embedded tissue with lymph node), PN (normal paraffin embedded tissue), PT (tumor paraffin embedded tissue), TM (tumor tissue), and TN (normal tissue). Depending on research needs, extraction is done from raw specimens, and the extraction type includes D (DNA), R (RNA), P (protein), etc. For a peripheral blood specimen received with serial number 1252 in year 2014, with a DNA sample then extracted from it, our assigned biobank number will be “BL-1252-14D.” This nomenclature system provides a clue about samples; however, patient confidentiality is protected as per current security regulations. To guarantee quality of samples, the biobank has ascertained standard policies via a [[quality management system]]. Specimens and samples are stored in liquid nitrogen, −80 °C, −20 °C, or 4 °C refrigerators depending on its type and requirement. Vials containing specimen or extracted sample are stored in an assigned area so that all aliquots can be retrieved in the LIMS from the defined physical location. | |||
===Evaluation of software features and performance efficiency=== | |||
We evaluated the efficiency of Biosearch System by performance testing using (i) Apache JMeter software, (ii) Create, Read, Update, and Delete (CRUD) function testing, and (ii) and a set of user feedback questionnaires. To verify the efficiency (i.e., speed, scalability, and stability) of the LIMS, a performance test was done through Apache JMeter by running different numbers of users (i.e., 500, 100, 50, 10, 1), with 50 loop and 10 ramp-up periods. CRUD function testing using different SQL statement was also used for performance testing. | |||
We also evaluated the LIMS through feedback provided by questionnaire concerning the LIMS' features and efficiency. Answers allowed for a response using a scale from 1 to 5 to specific questions such as: | |||
* Is it easy to reach the website application? | |||
* Does website application load quickly? | |||
* Are the fonts easy to read on various screen resolutions? | |||
* Is the color scheme appropriate and comfortable to the eye? | |||
* Is the content logically separated and appearing in an appropriate way? | |||
* Is the website application easy to use? | |||
* Is navigation easy? | |||
* Are all buttons (internal and external) valid and active? | |||
* Is the copy and paste feature allowed? | |||
* Is the autocomplete feature allowed? | |||
* Do clickable icons work smoothly on a single click? | |||
* Is the website application free from server-side errors? | |||
* Are you able to search, retrieve, and edit data (e.g., samples description, patient information, project details, etc.) easily? | |||
* Do you accurately receive an alert message for missing data? | |||
* Is data printed in an appropriate tabular format? | |||
* Is the help desk easy to contact for any issue in regards to the web-based application? | |||
| Line 73: | Line 103: | ||
'''CEGMR''': Center of Excellence in Genomic Medicine Research | '''CEGMR''': Center of Excellence in Genomic Medicine Research | ||
'''DEG''': | '''DEG''': differentially expressed gene | ||
'''LIMS''': laboratory information management system | '''LIMS''': laboratory information management system | ||
Revision as of 19:46, 4 July 2022
| Full article title | Development of Biosearch System for biobank management and storage of disease-associated genetic information |
|---|---|
| Journal | Journal of King Saud University - Science |
| Author(s) | Karim, Sajjad; Al-Kharraz, Mona; Mirza, Zeenat; Noureldin, Hend; Abusamara, Heba; Alganmi, Nofe; Merdad, Adnan; Jastaniah, Saddig; Kumar, Sudhir; Rasool, Mahmood; Abuzenadah, Adel; Al-Qahtani, Mohammed |
| Author affiliation(s) | King Abdulaziz University, Temple University |
| Primary contact | Email: skarim1 at kau dot edu dot sa |
| Year published | 2022 |
| Volume and issue | 34(2) |
| Article # | 101760 |
| DOI | 10.1016/j.jksus.2021.101760 |
| ISSN | 1018-3647 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.sciencedirect.com/science/article/pii/S1018364721004225 |
| Download | https://www.sciencedirect.com/science/article/pii/S1018364721004225/pdfft (PDF) |
Abstract
Objective: Databases and software are important to manage modern high-throughput laboratories and store clinical and genomic information for quality assurance. Commercial software is expensive, with proprietary code issues, while academic versions have adaptation issues. Our aim was to develop an adaptable in-house software system that can store specimen- and disease-associated genetic information in biobanks to facilitate translational research.
Methods: A prototype was designed per the research requirements, and computational tools were used to develop the software under three tiers, using Visual Basic and ASP.net for the presentation tier, SQL Server for the data tier, and Ajax and JavaScript for the business tier. We retrieved specimens from the biobank using this software and performed microarray-based transcriptomic analysis to detect differentially expressed genes (DEGs) with FC ±2 and P-value <0.05 in triple-negative breast cancer cases. The Ingenuity Pathway Analysis (IPA) tool was used to predict canonical molecular pathways associated with disease. Overall performance and utility of software was evaluated by Apache JMeter software, CRUD function testing, and a set of user feedback questionnaires.
Results: We developed Biosearch System, a web-based laboratory information management system (LIMS) enabling management of biobank samples (e.g., tissue, blood, FTTP slides) and their extracts (e.g., DNA, RNA, and proteins) with clinical and experimental details. The client satisfaction feedback was excellent, with a score of 4.7 out of 5. We identified a total of 1,181 DEGs, including both upregulated (IFI6, LEF1, FANCI, CASC5, PLXNA3, etc.) and down-regulated (ADH1B, LYVE1, ADH1C, ADH1B, ADIPOQ, PLIN1, LYVE1, etc.) genes in triple-negative breast cancer. Pathway analysis of DEGs revealed significant activation of interferon signaling (z-score 2.646) and kinetochore metaphase signaling pathway (z-score 2.138) in cancer.
Conclusion: Biosearch System is a user friendly LIMS for collection, storage, and retrieval of specimen and clinical information. It is secure, efficient, and convenient in sample tracking and data analysis. We illustrated its utility in transcriptomic study of breast cancer. Additionally, it can facilitate and speed up any genomic study and translational research publications.
Keywords: Biosearch System, LIMS, database, biobank, genomics, microarray, bioinformatics
Graphical abstract:
Introduction
Biobanks are established for long-term storage and conservation facility for biological specimens along with their demographic, clinical and experimental information to support scientific investigation using bioinformatics tools (Artene et al., 2013). That investigation, tied to high-throughput technological advancements in next-generation sequencing (NGS) and microarrays, has generated significant amounts of data at relatively low costs. (Diamandis, 2009, Fehniger and Marko-Varga, 2011, Glenn, 2011, Merdad et al., 2014) However, as the amount of biological specimens grows into the thousands, manual methods fail in the efficient handling of specimens and samples, and the risk of information being lost increases. Quality becomes another major concern with increasing volume of big data. As such, a robust software solution becomes necessary to effectively manage biobank and laboratory information and ensure the quality of results with approved ethical guidelines and personal integrity. ((ISBER), 2012, Bredenoord et al., 2011, Kang et al., 2013, Voegele et al., 2007) Software is helpful in managing the workflow cycle—comprising of collection, storage, analysis, and report generation—facilitating quick and easy retrieval of information and speeding up biomarker and therapeutic discoveries. (Melo et al., 2010)
The collection of a significant cohort size with crucial factors like number and type of samples, clinical information, pathological finding, follow-up data, etc. is a time-consuming process but is strongly recommended to facilitate comprehensive translational research. (Betsou et al., 2010, Hewitt, 2011, Huang et al., 2011, Riegman et al., 2008) Bioinformatics software aiding with these tasks is either available as commercial off-the-shelf (COTS), with proprietary code and high cost, or as academic or open-source, with complexity that is difficult to adopt within other laboratories. (Greely, 2007, Huang et al., 2011, Kauffmann and Cambon-Thomsen, 2008, Minamikumo, 2012, Prilusky et al., 2005) As such, we developed and implemented a software system to support biobank investigators in accessing all clinical and experimental disease-associated information required for basic and translational research. Herein, we discuss the application of our in-house laboratory information management system (LIMS), Biosearch System, for genomic analysis specifically for breast cancer transcriptomics, with further extensibility for any other diseases.
Materials and methods
Biosearch System was developed using a three-tier architecture model:
- Presentation tier: An interactive web browser for the end user’s computer
- Data tier: A SQL Server Management Studio that manages the storage and DB-server
- Business tier: A bridge between the other two tiers, facilitating the collection of data from the presentation tier and validating it before finally sending it to the data tier, and vice versa
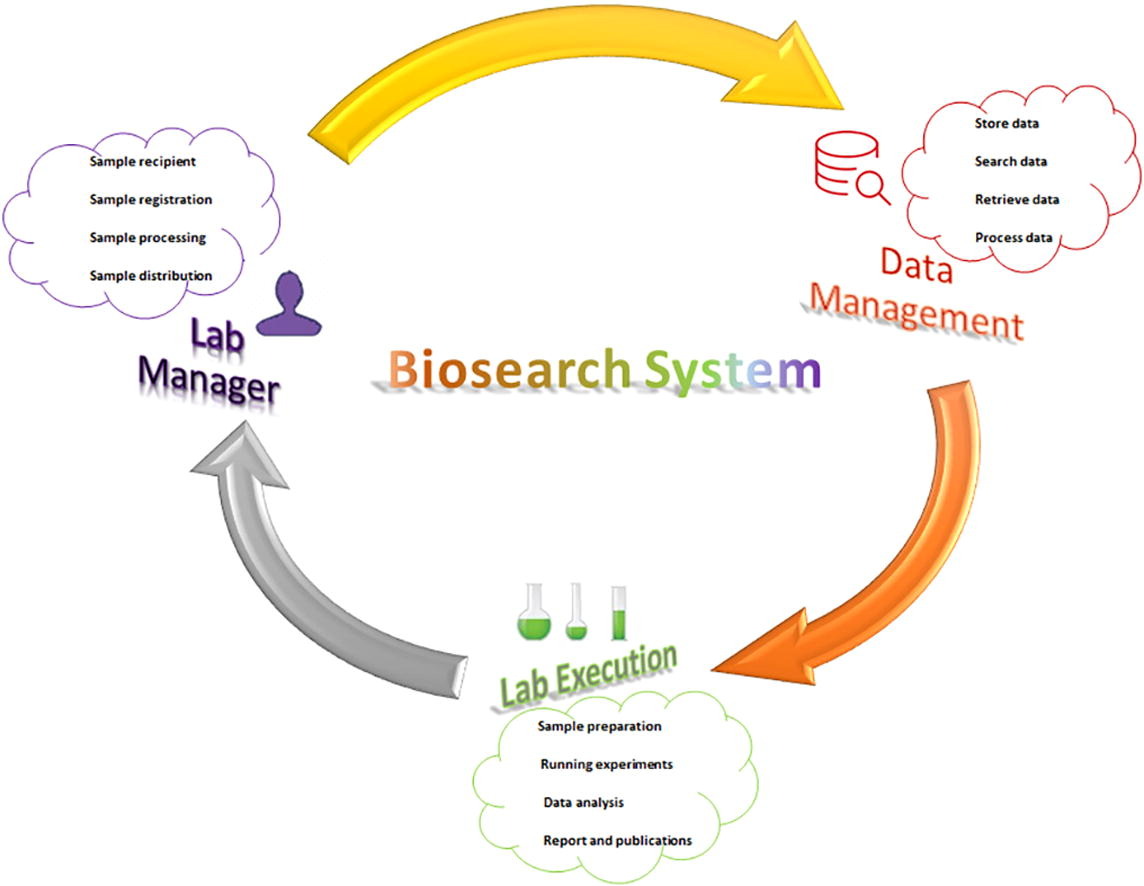
Biosearch System follows standard guidelines like legal requirements, standardized architecture, workflow, and dedicated staff members, as well as technical procedures to record and access clinical data, as described herein.
Legal requirements
The LIMS has been designed in accordance with the Saudi Arabian approved regulation for genomic medicine research. Patients were informed prior and written consents were taken by their doctors for their samples to be sent to the biobank for research. We provided consent forms to patients approved by the ethics committee. Additionally, the software stores data in coded format to hide the identity of patients during any research presentation and publications. In order to ensure bioethical safety, we followed Saudi's National Committee of Biological and Medical Ethics guidelines. (Royal decree No. M/59, dated 14/9/1431H – 24/8/2010)
Software and hardware architecture
We began with a prototype, an early model building process to test a proposed concept to enhance precision by system analysts and users. Database software design started by first designing a low fidelity prototype (paper-based prototype) and then designing a high-fidelity prototype (computer-based prototype). Collected information was categorized into logical groups or entities like "sample," "storage condition," "specimen request," "approval system," "project type," "diagnosis," and "disease type," and an entity-relationship diagram was built to show numerical relationship among different entities.
The LIMS itself was built on servers with following specifications: Windows Server 2012 R2 Enterprise Edition service packs 2, .NET 4.5, IIS 8.5, VS 2012, Microsoft SQL Server 2008 R2, ASP.NET, and AJAX Control Kit 4.5. A web browser was used for the graphical user interface (GUI). Database software was developed to support the high-performance hardware system and is compatible with common web browsers. To develop a robust and reliable LIMS, the following features were included:
- a web-based application for wide access to the database;
- a specimen and sample labelling mechanism before enrolling them into the database;
- security mechanisms that limit role-based access to authorized personnel; and
- a disaster management system to cope with any natural disaster.
System structure
To manage the laboratory's services, the software addresses the (i) control of specimen and sample data (i.e., add, edit, confirm transfer, retrieve, and search); (ii) viewing of the log book; (iii) display of a specimen or sample's status; (iv) viewing of box content; (v) addition of new entities, tests, categories, etc. (e.g., diagnostic tests, extractions, hospitals, sample types, projects); and (vi) request of a specimen or sample. Specimens and samples stored in the biobank are provided to researchers per project requirements and with proper justification and ethical approval. The requested sample is first checked by biobank staff then forwarded to the lab manager, before finally going to the director for final approval. Researchers can track the processing steps while the software updates the decision status by email. After delivery of approved specimens, requisite volume is automatically subtracted from biobank stock.
Database structure
A normal relational database is used for the system. Information is stored in specific tables like the SampleInfo table, which contains the sample's information, the ProductInfo table, which contains the type of sample information (e.g., DNA, RNA), the PatientInfo table, which contains a patient's related information, the SampleStorage table, which contains the storage location information (e.g., refrigerators, shelf, box container), and the UsersInfo table, which contains user-related information. The type of relation is one-to-many (one patient::multi-samples, one sample::multi-products, one refrigerator::multi-samples, etc.).
Workflow and biobank organizational structure
The software can only be used with an authorized username and password, and after successful login on a web-based client PC, users are allowed to add new data, update/edit data, follow up with patients, and retrieve data for analysis and interpretation. Biosearch System also manages the storage, quality, quantity, distribution, and maintenance of specimens (e.g., tissue, blood, serum, etc.) and its derivatives (e.g., DNA, RNA, protein, plasma, etc.). Per the existing CEGMR biobank organizational structure, assigned users have different level of rights for using the software. Researchers are end users of the biobank; they request samples based on their active research projects. Dedicated biobank staff examine the request and update the status, handing it off to the supervisor, who approves or rejects the request with valid reason. However, the final decision is made by the director, who has ultimate authority to reconsider the request or reverse the supervisors' decision. Once approved by the director, the biobank delivers the requested items within a week. Reminders for pending tasks are emailed to relevant staff until the final decision is made. This system enables researchers full access to the biobank information by allowing for the management of specimens and samples, the notification for request approval or rejection, and provision of the overall summary of the biobank’s inventory to researchers.
Technical procedure for linking clinical information to biobank specimens
The LIMS manages all the relevant information for all deposited patients’ specimens and samples for systematic clinical research. We use a medical record number (MRN) as the primary key and a biobank number as the secondary key to connect two sections of the database. Patients’ samples are stored at the biobank with a unique allocated biobank number, which consists of 10 digits formatted as sample type (XX), serial number (XXXX), year (XX), and extraction type (XX). Sample types in our database includes AM (amniotic fluid), BL (blood), BO (bone marrow), CO (cord blood), CS (cervix swab), LN (lymph node), PC (product of conception), PL (paraffin embedded tissue with lymph node), PN (normal paraffin embedded tissue), PT (tumor paraffin embedded tissue), TM (tumor tissue), and TN (normal tissue). Depending on research needs, extraction is done from raw specimens, and the extraction type includes D (DNA), R (RNA), P (protein), etc. For a peripheral blood specimen received with serial number 1252 in year 2014, with a DNA sample then extracted from it, our assigned biobank number will be “BL-1252-14D.” This nomenclature system provides a clue about samples; however, patient confidentiality is protected as per current security regulations. To guarantee quality of samples, the biobank has ascertained standard policies via a quality management system. Specimens and samples are stored in liquid nitrogen, −80 °C, −20 °C, or 4 °C refrigerators depending on its type and requirement. Vials containing specimen or extracted sample are stored in an assigned area so that all aliquots can be retrieved in the LIMS from the defined physical location.
Evaluation of software features and performance efficiency
We evaluated the efficiency of Biosearch System by performance testing using (i) Apache JMeter software, (ii) Create, Read, Update, and Delete (CRUD) function testing, and (ii) and a set of user feedback questionnaires. To verify the efficiency (i.e., speed, scalability, and stability) of the LIMS, a performance test was done through Apache JMeter by running different numbers of users (i.e., 500, 100, 50, 10, 1), with 50 loop and 10 ramp-up periods. CRUD function testing using different SQL statement was also used for performance testing.
We also evaluated the LIMS through feedback provided by questionnaire concerning the LIMS' features and efficiency. Answers allowed for a response using a scale from 1 to 5 to specific questions such as:
- Is it easy to reach the website application?
- Does website application load quickly?
- Are the fonts easy to read on various screen resolutions?
- Is the color scheme appropriate and comfortable to the eye?
- Is the content logically separated and appearing in an appropriate way?
- Is the website application easy to use?
- Is navigation easy?
- Are all buttons (internal and external) valid and active?
- Is the copy and paste feature allowed?
- Is the autocomplete feature allowed?
- Do clickable icons work smoothly on a single click?
- Is the website application free from server-side errors?
- Are you able to search, retrieve, and edit data (e.g., samples description, patient information, project details, etc.) easily?
- Do you accurately receive an alert message for missing data?
- Is data printed in an appropriate tabular format?
- Is the help desk easy to contact for any issue in regards to the web-based application?
Abbreviations, acronyms, and initialisms
CEGMR: Center of Excellence in Genomic Medicine Research
DEG: differentially expressed gene
LIMS: laboratory information management system
MRN: medical record number
References
Notes
This presentation is faithful to the original, with minor changes to presentation; grammar and spelling required more cleanup for improved readability. In some cases important information was missing from the references, and that information was added.