Journal:From biobank and data silos into a data commons: Convergence to support translational medicine
| Full article title | From biobank and data silos into a data commons: Convergence to support translational medicine |
|---|---|
| Journal | Journal of Translational Medicine |
| Author(s) | Asiimwe, Rebecca; Lam, Stephanie; Leung, Samuel; Wang, Shanzhao; Wan, Rachel; Tinker, Anna; McAlpine, Jessica N.; Woo, Michelle M.M.; Huntsman, David G.; Talhouk, Aline |
| Author affiliation(s) | BC Cancer Research Centre, BC Children’s Hospital Research Institute, University of British Columbia, OVCARE |
| Primary contact | Email: a dot talhouk at ubc dot ca |
| Year published | 2021 |
| Volume and issue | 19 |
| Article # | 493 |
| DOI | 10.1186/s12967-021-03147-z |
| ISSN | 1479-5876 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-021-03147-z |
| Download | https://translational-medicine.biomedcentral.com/track/pdf/10.1186/s12967-021-03147-z.pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Background: To drive translational medicine, modern day biobanks need to integrate with other sources of data (e.g., clinical, genomics) to support novel data-intensive research. Currently, vast amounts of research and clinical data remain in silos, held and managed by individual researchers, operating under different standards and governance structures; such a framework impedes sharing and effective use of data.
In this article, we describe the journey of British Columbia’s Gynecological Cancer Research Program (OVCARE) in moving a traditional tumor biobank, outcomes unit, and a collection of data silos into an integrated data commons to support data standardization and resource sharing under collaborative governance, as a means of providing the gynecologic cancer research community in British Columbia access to tissue samples and associated clinical and molecular data from thousands of patients.
Results: Through several engagements with stakeholders from various research institutions within our research community, we identified priorities and assessed infrastructure needs required to optimize and support data collections, storage, and sharing, under three main research domains: (1) biospecimen collections, (2) molecular and genomics data, and (3) clinical data. We further built a governance model and a resource portal to implement protocols and standard operating procedures (SOPs) for seamless collections management and governance of interoperable data, making genomic and clinical data available to the broader research community.
Conclusions: Proper infrastructures for data collection, sharing, and governance is a translational research imperative. We have consolidated our data holdings into a data commons, along with standardized operating procedures to meet research and ethics requirements of the gynecologic cancer community in British Columbia. The developed infrastructure brings together diverse data and computing frameworks, as well as tools and applications for managing, analyzing, and sharing data. Our data commons bridges data access gaps and barriers to precision medicine and approaches for diagnostics, treatment, and prevention of gynecological cancers by providing access to large datasets required for data-intensive science.
Keywords: biobanks, biospecimens, biobank technologies. precision medicine, data commons, laboratory information management systems, LIMS, federated systems, data governance
Background
The collection, storage, management, and distribution of human biospecimens for diagnostic pathology [1,2,3] can be traced as far back as the 1900s. [3] To meet research needs in the postgenomic era, modern day biorepositories [4] support scientists to derive disease-specific insights [5] by aiding the investigation of genetic underpinnings [6,7,8], elucidating etiology, and evaluating disease progression and therapeutic response; they are the backbone of precision medicine [9, 10], as well as biomedical and translational research. [1, 2, 11]
The last decade has seen advances in biotechnology such as next-generation sequencing (NGS), and the emergence of “omics” techniques for precision medicine (e.g., genomics, transcriptomics, proteomics, metabolomics, and epigenomics). These innovations coincided with breakthroughs in computing, artificial intelligence (AI), and analytics, enabling discrimination between disease with greater precision. [12] This has created an unprecedented demand for high-quality biospecimens and associated data, including clinical, molecular, imaging, and other types of data generated during research. [11] Innovations in database cloud storage and computing infrastructures to support data-intensive science have further contributed to revolutionizing resources available to address modern research needs. [13, 14] As federated models for aggregating data and biomaterials have emerged as favored approaches for identifying enough patients with specific clinical or molecular features, the importance of interoperability between biobanks and related databases has been accentuated. [4, 7, 15] Specimen collections have become virtual [13], flexible, and interoperable, hosted on internationally harmonized infrastructures [7] and optimized for secondary research. [7, 13] Present-day research environments and needs have led to the development and implementation of data commons [16, 17], bringing together within a research community diverse data and computing infrastructures, as well as tools and applications for managing, analyzing, and sharing interoperable data. This has created an opportunity to maximize collaborations and to extend the value generated from primary data collection. [18]
In 2016, as part of British Colombia’s multidisciplinary gynecological cancer research team (OVCARE), we undertook a comprehensive review of the landscape of the data assets available within our local research environment and assessed the infrastructure needs required to support data storage and sharing within our research community. Herein, we describe the roadmap undertaken for the creation of a data commons, transforming a traditional tumor biobank and a collection of data silos into an integrated and comprehensive infrastructure to support the current and future research needs of an expanding team.
Results
Matching technical solutions to research needs
OVCARE started in 2000 as an initiative between the British Columbia Cancer Agency, the University of British Columbia, and the Vancouver Coastal Health Research Institute to accelerate research discoveries and their translation to the clinical settings, as well as improve the lives of women with ovarian cancer or those at risk. Today, OVCARE is an internationally recognized multidisciplinary team of physicians and scientists who are breaking new ground in improving diagnosis, prevention, and treatment of all gynecological cancers. [19,20,21,22,23,24,25,26,27]
OVCARE’s research has been powered by the gynecological tumor bank and the Cheryl Brown Gynecological Cancers Outcomes Unit. Through the course of research, a plethora of molecular and genomic data was historically held by researchers that generated them. Similarly, clinical data from chart reviews are obtained to support clinical studies and held with clinicians. These data were in incompatible formats that needed significant manual manipulation and curation to be integrated. Moreover, each collection was governed by different ethics agreements that restricted the use of data and kept it in silos. This was becoming a barrier to novel data-intensive research, requiring the integration of multiple data sources; undertaking such projects was challenging, time consuming, and prone to errors. The OVCARE leadership had recognized that current research needs were not being met through existing infrastructure.
A broad stakeholder engagement effort in 2016 kicked off, with the objective to work with researchers, clinicians, scientists, and technicians at various institutions, to map out a collective future vision, identifying research needs and re-thinking present infrastructure. Engagements with key stakeholders identified research priorities, which were expanded into a list of fundamental requirements (Table 1) relevant to the collection and optimization of biospecimen, clinical, and molecular/genomics data, as well as a governance model of the resulting infrastructure. In addition to generating efficiency, limiting errors and honoring patient consent, fundamental research requirements included the maximization of secondary use of data, which enables data collected for one purpose to be reused in a completely different context. For example, chemotherapy drugs dispensed at our pharmacy are collected for administrative purposes (billing) but can also be used to link with patient phenotype, genotype, and outcome to investigate which patients benefit from these therapies more than others. Another important need was to generate novel research hypotheses by considering simultaneously various data that could never before be considered at the same time. Patterns that may not have been obvious previously may emerge to drive future innovative research. Another important need was to use translational studies to help inform patient care, as well as use data generated from patient care to ask new research questions, with the goal of continuously trying to better fill gaps in understanding of disease etiology and progression. In upcoming sections, we further describe more of these requirements in greater detail.
| ||||||||||||||||||||||||||||||||||||||||||||
Biospecimen collection
OVCARE employs two models for biospecimen recruitment. The first is a general banking model, with broad scientific aims, and where specimens are obtained from consented participants and stored until needed. The second is a study-based banking model, where participants are recruited to address specific study aims, with a pre-defined protocol and pre-planned specimen collection. To accommodate both approaches, the biorepository infrastructure needed to manage accrual of specimens in a patient-centric approach, retain the context of the patient’s clinical history, and support basic biospecimen collection, storage, and distribution across multiple studies at different sites, under both recruitment models. This includes inventory control, the ability to track sample availability and location, as well as track generated derivatives (e.g., xenografts and organoids). The infrastructure needed to be adaptable to changing needs between studies and projects, as well as over time, with the ability to preserve the natural history of the data. Access control that varied for different user groups was a critical feature, enabling adherence to regulatory requirements and health research best practices. Data security, de-identification of specimens, and tracking of consent were also important for the same reasons, in addition to the need to operate and manage the biorepository with minimal support from institutional and research IT.
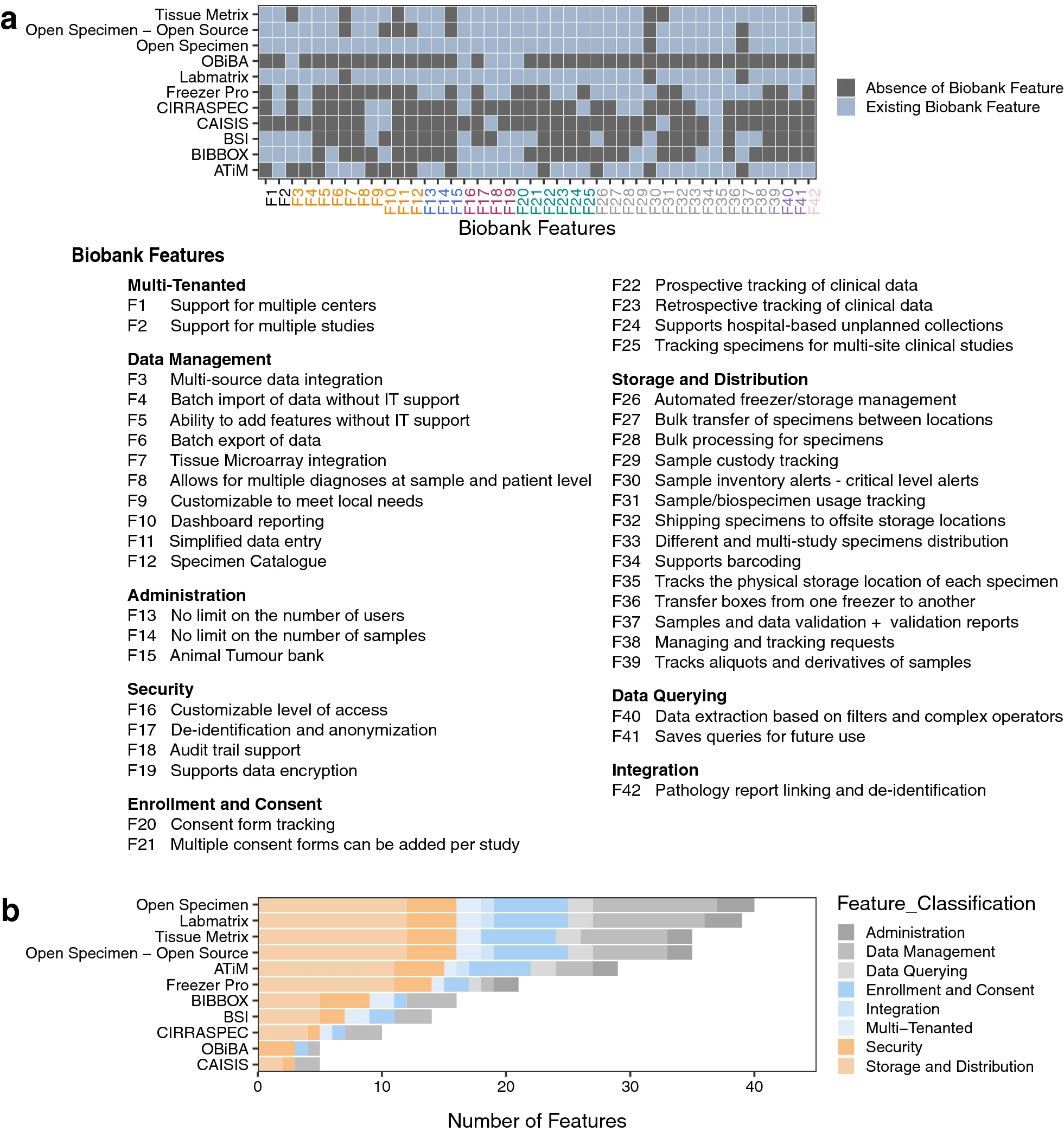
We compiled a comprehensive list of requirements (Additional file 2: Table S1) from our stakeholder meetings, and we used it to guide our scan of the landscape of existing laboratory information management systems (LIMS) (Additional file 2: Table S2—S11, and Fig. 1). This resulted in the identification of OpenSpecimen [28], a LIMS based on caTissue [29], a mature system with over 15 years of use by the research community. OpenSpecimen addressed more requirements from our list in comparison to other options we considered. It is an open-source software solution with commercial support, in use by over 70 biobanks across 20 countries. The commercial support ensures ongoing software testing, updating, and continuous improvement. This is in addition to the availability of technical support, and access to a community of experienced users through active forums.
|
In this LIMS, biospecimens can be processed individually or in bulk, with rapid barcode-based scanning available to enter information on multiple patient samples at once. This enabled high-throughput processing and efficient migration from our legacy LIMS. Options for data annotation and storage management allowed us to optimize specimen storage, a costly resource in our research community (e.g., − 80 freezers). [29]
The OpenSpecimen LIMS enabled customization of data entry forms via a graphical user interface (GUI, or web interface) to match study-specific needs without requiring software development. The platform met most of our IT requirements as it supported role-based access control and provided an audit trail of every user operation. [30] The system was also easy to use, with graphics-based queries that enable searching for stored data about participants, biospecimens, or projects without requiring any programming, making the moderately complex queries accessible to most users. Queries could also be performed via REST API (Representational State Transfer Application Programming Interface) using a SQL (Structured Query Language)-like query language. This facilitated automation of data downloads for analytics pipelines through the incorporation of query scripts.
The system enables standalone plugins through a software development kit. These plugins can be made publicly available to the community. For example, the tissue microarray (TMA) plugin can manage TMAs on OpenSpecimen by linking to donor blocks and describing details of experiments done on the different slices of the TMA blocks. Finally, interoperability with other systems was important to expand linkage within the data commons. The vendor provides integration with electronic data capture applications (REDCap, OpenClinica), electronic medical record (EMR) systems (EPIC, Velos), and pathology systems (CoPath, Cerner, Aperio), as well as systems using Health Level 7 (HL7) messages, a capability which can further support inclusion of participant and biospecimen information from distributed systems.
Molecular and genomics data
Various molecular and genomics data are generated through the course of research, including next-generation sequencing, proteomics, gene expression, targeted sequencing, and immunohistochemical data. These data are primarily generated to answer specific research hypotheses and were supported by public, government, and philanthropic funds, with an implicit obligation to minimize duplication of efforts and to optimize their secondary use in later research. The ability to consider all this data simultaneously can uncover novel patterns, trends, and unknown correlations. This may prompt new hypotheses and spark new insights into novel research directions. To achieve this level of integration, we would need to track which analytical assay was performed on which samples and link back to those data. To facilitate the interrogation of this complex data, an exploration tool was needed to visualize resulting multidimensional datasets and simultaneously investigate molecular profiles and clinical attributes.
We adopted the cBio Cancer Genomics Portal [31], one of the most recommended and widely used [32,33,34,35,36] pan-cancer analytics web tools to facilitate interactive exploration, mining, analysis, and visualization of multidimensional datasets derived from tumor samples collected from various cancer studies. [31, 37] Developed at the Memorial Sloan Kettering Cancer Center (MSK), this platform is used by large cancer genomic studies (TCGA [38], TARGET [39]), and publicly available data can be downloaded and queried alongside our own collections.
The cBio Cancer Genomics Portal enables the collection of various genomic data on each tumor sample, including non-synonymous mutations, copy-number alterations (CNAs), mRNA and microRNA expression data, DNA methylation data, protein data, and phosphoprotein level data. [31] Each of these data types is integrated and stored at the gene level to allow investigators to probe for the presence of specific biological events (e.g., gene mutations, deletions, amplifications, and expression levels in each sample) [37], and compare discrete genomic events and patterns across samples and across multiple integrated data types. [31] Stored gene-level data is integrated with de-identified clinical data to probe patient clinical outcomes to support the development or testing of hypotheses on frequently altered genes in specific cancers. [31, 37] In addition, it enables the investigation of the prognostic roles of certain genes in gynecological and other cancers [34], correlations between mutations, expression profiles, clinicopathological features, and potential diagnostic and therapeutic targets for certain cancer types.
Clinical data
Clinical data at OVCARE are obtained and collected for the purpose of evaluation of patient outcomes and improvement of the quality of patient care and research. Some of these data were historically managed by the Cheryl Brown Outcomes Unit for the purpose of outcomes research on ovarian cancer patients referred to BC Cancer, the provincial tertiary cancer center. The BC Cancer Registry provided the Cheryl Brown Outcomes Unit regular data updates, such as the identification of patients with cancer and their vital statistics, which were supplemented by exhaustive chart reviews. In addition to the Cheryl Brown Outcomes Unit, clinicians often conducted chart reviews for other clinical studies; the resulting data was held separately. In 2016, the scope of data collection at the Cheryl Brown Outcomes Unit was limited to ovarian cancer and did not take full advantage of other available data. Collecting clinical data was resource-intensive and the effort needed was not sustainable in the long run. Moreover, the mandate of the Cheryl Brown Outcomes Unit expanded to enable OVCARE’s researchers to study all gynecological cancers in the province of BC, especially those cancers that do not require referral to a cancer center (e.g., in BC, up to 50% of patients with endometrial cancer are treated by gynecologists in their communities). Thus, an important priority for the team was to create efficiencies in clinical data collection and to standardize, integrate, and link all gynecologic cancer clinical data from various sources and consolidate clinical data in a single database. This would allow researchers to understand what clinical data is already available, thereby streamlining their own data collection strategies which would, in turn, directly contribute to a master database. To maximize the re-use of clinical data, standardization of ontologies across projects was needed, as well as the creation of infrastructure to serve as permanent storage with an easy-to-use data collection interface adaptable to fit the needs of various research projects. This would allow standardization of data collection, to the greatest extent possible, and minimization of errors. Consequently, this would improve the overall quality of data, maximize interoperability and reusability, and optimize data analysis. Management of sensitive clinical data requires security, privacy, and the use of tools and technology with institutional approval. We also needed rigorous security and privacy measures, and comprehensive audit trails for tracking data manipulation, exports, and downloads for both single and multi-centered research studies, including tracking data access.
To support OVCARE’s clinical data requirements, we adopted Research Electronic Data Capture (REDCap), a widely used, free and flexible web-based application [40, 41] developed at Vanderbilt University for clinical and translational research. It is one of the most popular research electronic data systems implemented in 141 countries by over 1,000,000 [42] studies, including our institutions. REDCap’s flexible design supports permanent database collections, which can be augmented by both patient/study-centric surveys or data collection forms, and includes a rich set of modules that support today’s diverse and multi-scaled biomedical research operations. [41]
Governance structure
To manage the various integrated datasets (biospecimen, molecular, genomic, and clinical data), we needed to ensure proper governance, protocols, and standard operating procedures (SOPs) to support data sharing, streamline data requests and inquiries, undertake scientific review or requests, and ensure availability of ethics approval. We envisioned a single portal application for all requests and queries, with a backend database keeping track of details of requesting researchers, description of projects, and required resources, as well as their associated ethics application and certificates of approval. This infrastructure would facilitate compliance with ethics and maintain a log of all activities.
We adopted Oracle Corporation's Oracle Application Express (APEX) [43] to develop this portal application. Already available at our institution, APEX, is a low-code, data-driven platform for rapid development and deployment of scalable and secure web applications. Applications are implemented in a preconfigured environment; all development was done through a web interface that is mostly GUI–based. The middle-tier functions of the web application software stack, such as parsing Hypertext Transfer Protocol (HTTP) requests and session management, are fully automated, and all operational aspects of the system (data backup, software patches and updates) are managed by institutional IT.
Implementation
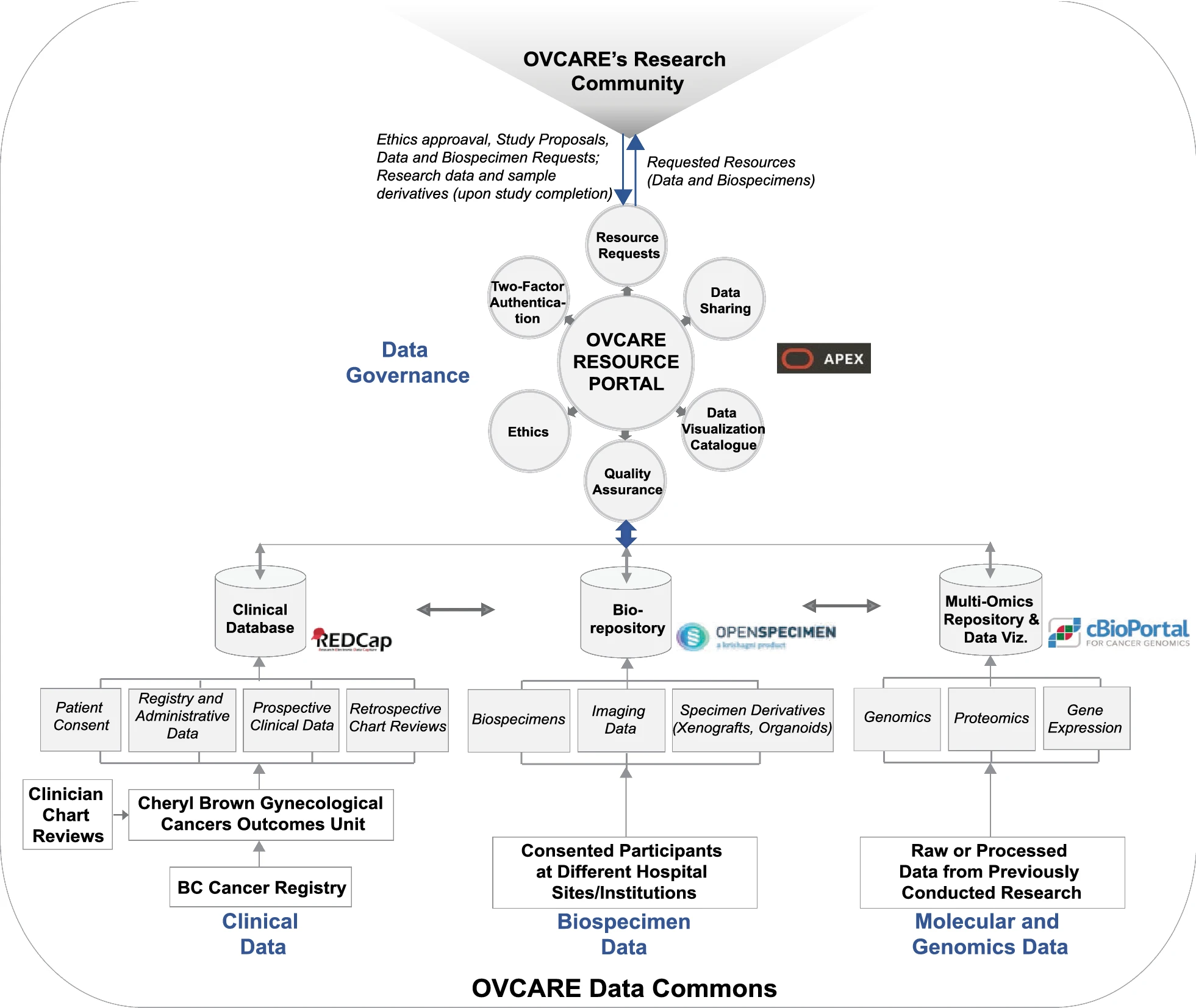
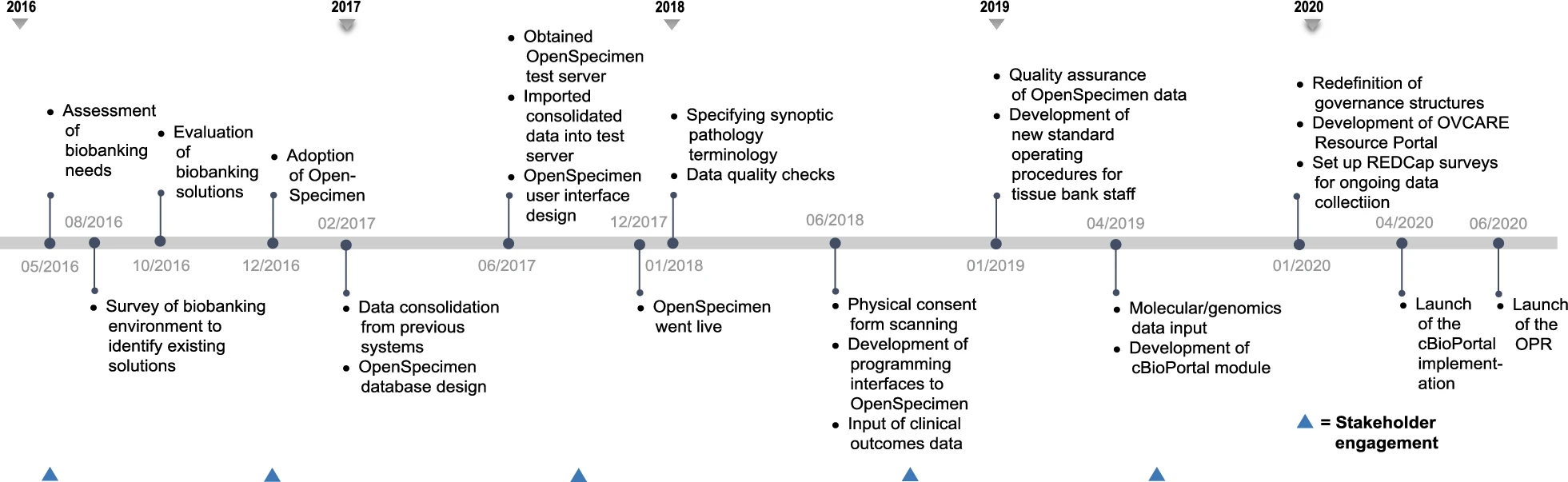
The various components of the data commons infrastructure and software identified to meet the domain-specific needs described in the previous section are illustrated in Fig 2. This infrastructure is implemented behind institutional firewalls, with only the resource portal accessible through the world wide web. The path to implementing this infrastructure was not linear and continues to evolve, despite the linear timeline (presented in Fig. 3).
|
|
In early 2017, we completed a survey of existing biobanking solutions to select one that provided the best fit to our needs at that time. In June 2017, a test server was obtained to run local instances of the selected LIMS, OpenSpecimen, to conduct functionality, integration, and unit testing of all components of this software. This enabled us to evaluate OpenSpecimen's features firsthand and to determine the required resources to operate the infrastructure with optimal performance in our current computing and research environment. We tested for performance and evaluated operation workflows by diverse types of users, both technical and nontechnical, to perform daily biobanking activities. We fully adopted OpenSpecimen in December of 2017. Following this migration, we worked with researchers to gather available genomic datasets and link their availability to the respective biospecimen in OpenSpecimen as well as indicate where data are held. As we continue to expand this resource, we will add availability of images of pathology slides, associated with each tumor block and link to them. To prototype the cBio Cancer Genomics Portal integration, we gathered molecular data for one ovarian cancer subtype, collected from prior studies which were integrated with specimen availability and key clinical outcomes in cBio Cancer Genomics Portal, using specimen ID. We recently launched this prototype and it is currently under evaluation.
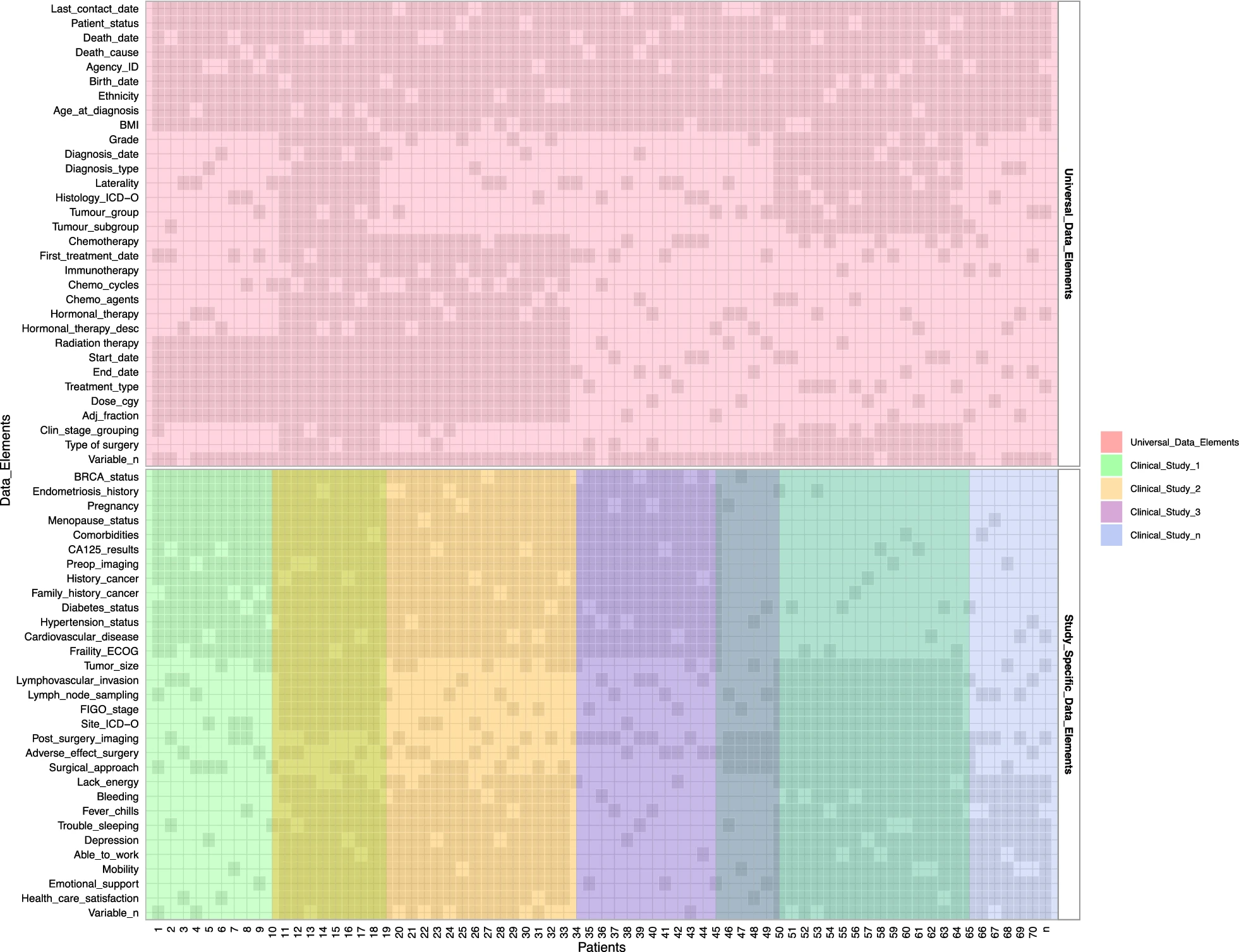
For clinical data, we expanded the mandate of the Cheryl Brown Outcomes Unit to include clinical and outcome data on all gynecological cancer patients diagnosed in British Columbia. We also obtained ethics approval to permanently retain clinical and outcomes data from all clinical studies in our group. We maximized data we can receive from administrative sources, such as the BC Cancer Registry, as this provides access to clinical data for all patients and minimizes the need for broad chart reviews (Fig. 4). We included elements, such as the date of diagnosis, date of last clinical appointment, vital statistics, International Classification of Diseases (ICD)-10 morphology codes, tumour stage, and grade. We are presently investigating additional data, such as systemic therapy (chemotherapy and radiation therapy received). The second step of clinical data integration involved adding clinical studies with chart reviews. To enable that, we needed to map different data elements to unique concepts. This further facilitated the identification of variables that are of greatest interest to researchers in our group. We then developed consistent data definitions, standards, and semantics for each data element to ensure that all data can be integrated within the data commons. Future data collection will consult these data standards to ensure prospectively harmonized clinical data.
|
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.
[[Category:LIMSwiki journal articles (added in 2019)]