Journal:Information management for enabling systems medicine
| Full article title | Information management for enabling systems medicine |
|---|---|
| Journal | Current Directions in Biomedical Engineering |
| Author(s) | Ganzinger, Matthias; Knaup, Petra |
| Author affiliation(s) | Heidelberg University's Institute of Medical Biometry and Informatics |
| Primary contact | Email: matthias dot ganzinger at med dot uni-heidelberg dot de |
| Year published | 2017 |
| Volume and issue | 3 (2) |
| Page(s) | 0105 |
| DOI | 10.1515/cdbme-2017-0105 |
| ISSN | 2364-5504 |
| Distribution license | Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International |
| Website | https://www.degruyter.com/view/j/cdbme.2017.3.issue-2/cdbme-2017-0105/cdbme-2017-0105.xml |
| Download | https://www.degruyter.com/downloadpdf/j/cdbme.2017.3.issue-2/cdbme-2017-0105/cdbme-2017-0105.xml (PDF) |
Abstract
Systems medicine is a data-oriented approach in research and clinical practice to support the study and treatment of complex diseases. It relies on well-defined information management processes providing comprehensive and up-to-date information as the basis for electronic decision support. The authors suggest a three-layer information technology (IT) architecture for systems medicine and a cyclic data management approach, including a knowledge base that is dynamically updated by extract, transform, and load (ETL) procedures. Decision support is suggested as case-based and rule-based components. Results are presented via a user interface to acknowledging clinical requirements in terms of time and complexity. The systems medicine application was implemented as a prototype.
Keywords: systems medicine, information management, decision support systems
Introduction
Systems medicine is a current approach to aid physicians and researchers in the treatment and investigation of complex diseases. According to the definition of the European Commission, “‘Systems medicine‘ is the application of systems biology approaches to medical research and medical practice. Its objective is to integrate a variety of biological/medical data at all relevant levels of cellular organization using the power of computational and mathematical modelling, to enable understanding of the pathophysiological mechanisms, prognosis, diagnosis and treatment of disease.“[1] Consequently, the management of data is of great importance for systems medicine in research as well as clinical practice. Typically, data of different sources such as electronic health record systems, clinical research databases, or biomedical knowledge representations like ontologies have to be reviewed and prepared. The most prevalent data sources in systems medicine research projects are omics data and clinical data.[2]
Due to the comprehensive approach of systems medicine, neither disease-specific knowledge nor clinical data can be considered static. Thus, we suggest understanding information management for systems medicine as a dynamic process that evolves over time and leads to cyclic updates of the knowledge and data repositories behind the corresponding information technology (IT) system.
Further challenges arise from the broad availability of so-called omics data. This class of data — for example RNA microarray data — is characterized by a huge amount of attributes per sample that is often disproportional to the number of available cases. Currently, specific data preparation pipelines using statistical approaches like feature selection are necessary to make these data accessible for decision support solutions.[3]
Methods
For successful information management in the context of systems medicine, it is useful to distinguish between the IT architecture and the data management process. The architecture depends on the requirements of a specific systems medicine application. As a generic high-level architecture we propose a three-layer model[4]:
1. Data representation: Data and knowledge from different sources have to be prepared and made available for use in systems medicine. This includes data harmonization, transformation, and storage.
2. Decision support: Data and knowledge from layer 1 are processed by applying decision support approaches like case-based reasoning (CBR), deductive classifiers (rules-based), or systems biology models. Depending on the context, such components can be combined into hybrid systems.
3. User interface: Systems medicine applications should be designed to assist and not replace human decisions. Consequently, the user interface for such an application must be carefully designed to support well-informed, reproducible clinical decisions in an appropriate time frame.
The complexity of the data management process depends on the level of heterogeneity prevalent in the data sources. To achieve sufficient case numbers, it is often necessary to combine data on the same entity types from different sources. For example, hospitals may decide to collaborate and share clinical data on a specific disease area to build a joint systems medicine application with a higher number of cases and therefore greater statistical power (multi-center approach).
In most cases, clinical documentation will not be based on identical specifications. Thus, in a harmonization step, data definitions have to be evaluated for each attribute, both on a syntactic and semantic level. The resulting common data definition should be implemented into an automated extract, transform, and load (ETL) process to facilitate repeated loading of data to keep an up-to-date decision support system.[5][6]
Results
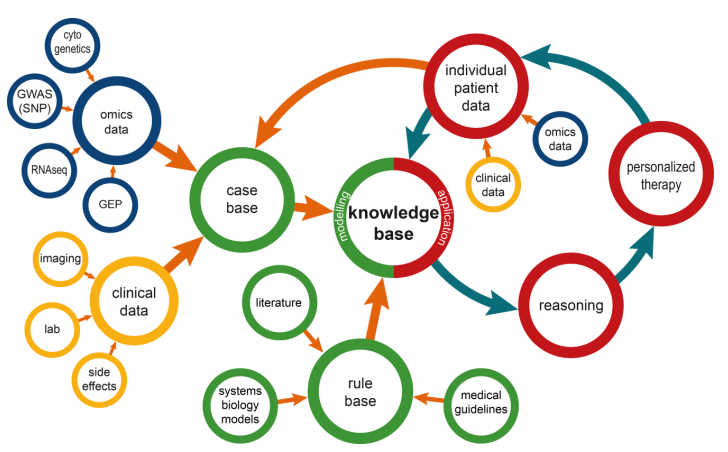
An overview of the resulting information management modelis shown in Figure 1. In the following paragraphs the elements of this model are described in detail.
|
Knowledge base
The core concept of the model is the knowledge base, which contains patient and disease related data, as well as formally represented knowledge. As such, it forms a systems medicine model in a broader sense. Specifically, the knowledge base is comprised of a case base and a rule base.
Case base
The case base covers information on the available experience in treating patients with a specific disease. Typically, such information is organized as case descriptions. Each case is described by a harmonized set of attributes covering clinical data, omics data, and others. The case base can be used for various research purposes like data mining or construction of systems biology models. In addition, its cases can be used for decision support directly by using the concept of patient similarity.[7] In the field of clinical artificial intelligence systems, patient similarity has been the subject of research for many years, most notably in case-based reasoning.[8]
References
- ↑ Auffray, C.; Balling, R.; Bensen, M. et al. (15 June 2010). "From Systems Biology to Systems Medicine". In Kyriakopoulou, C.; Mulligan, B. (PDF). European Commission. http://ec.europa.eu/research/health/pdf/systems-medicine-workshop-report_en.pdf.
- ↑ Gietzelt, M.; Löpprich, M.; Karmen, C. et al. (2016). "Models and Data Sources Used in Systems Medicine: A Systematic Literature Review". Methods of Information in Medicine 55 (2): 107–13. doi:10.3414/ME15-01-0151. PMID 26846174.
- ↑ Anaissi, A.; Goyal, M.; Catchpoole, D.R. et al. (2015). "Case-based retrieval framework for gene expression data". Cancer Informatics 14: 21–31. doi:10.4137/CIN.S22371. PMC PMC4368049. PMID 25861214. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4368049.
- ↑ Ganzinger, M.; Gietzelt, M.; Karmen, C. et al. (2015). "An IT Architecture for Systems Medicine". Studies in Health Technology and Informatics 210: 185-9. PMID 25991127.
- ↑ Firnkorn, D.; Ganzinger, M.; Muley, T. et al. (2015). "A Generic Data Harmonization Process for Cross-linked Research and Network Interaction. Construction and Application for the Lung Cancer Phenotype Database of the German Center for Lung Research". Methods of Information in Medicine 54 (5): 455-60. doi:10.3414/ME14-02-0030. PMID 26394900.
- ↑ Vassiliadis, P. (2009). "A Survey of Extract–Transform–Load Technology". International Journal of Data Warehousing and Mining 5 (3): 27. doi:10.4018/jdwm.2009070101.
- ↑ Brown, S.A. (2016). "Patient Similarity: Emerging Concepts in Systems and Precision Medicine". Frontiers in Physiology 7: 561. doi:10.3389/fphys.2016.00561. PMC PMC5121278. PMID 27932992. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5121278.
- ↑ Aamodt, A.; Plaza, E. (1994). "Case-based reasoning: foundational issues, methodological variations, and system approaches". AI Communications 7 (1): 39–59.
Notes
This presentation is faithful to the original, with only a few minor changes to grammar, spelling, and presentation, including the addition of PMCID and DOI when they were missing from the original reference.