Journal:Laboratory testing methods for novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
| Full article title | Laboratory testing methods for novel Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2) |
|---|---|
| Journal | Frontiers in Cell and Developmental Biology |
| Author(s) | D'Cruz, Roshan J.; Currier, Arthur W.; Sampson, Valerie B. |
| Author affiliation(s) | Nemours/Alfred I. duPont Hospital for Children |
| Primary contact | Email: valerie dot sampson at nemours dot org |
| Editors | LaBarge, Mark A. |
| Year published | 2020 |
| Volume and issue | 8 |
| Article # | 468 |
| DOI | 10.3389/fcell.2020.00468 |
| ISSN | 2296-634X |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.frontiersin.org/articles/10.3389/fcell.2020.00468/full |
| Download | https://www.frontiersin.org/articles/10.3389/fcell.2020.00468/pdf (PDF) |
Abstract
Following the first reports of coronavirus disease 2019 (COVID-19) by China to the World Health Organization (WHO) on December 31, 2019, more than 4,302,774 cases of infection by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus have been reported by authorities in 212 countries and territories as of May 12, 2020. The outbreak and spread of COVID-19 worldwide highlights the critical need for developing rapid and accurate diagnostic testing methods for emerging human coronavirus (CoV) infections. Testing is crucial to tracking the spread of disease during a pandemic, and to swiftly permitting public health interventions, including isolation, quarantine, and appropriate clinical management of afflicted individuals. The key components of viral diagnostic tests are (1) collection of the appropriate sample (blood, nasal swab, and throat swab); (2) availability of the genetic and proteomic sequences of the novel virus for analysis; and (3) rapid and accurate laboratory testing methods. The current gold standard for the molecular diagnosis of SARS-CoV-2 infection is the real-time reverse transcription polymerase chain reaction (qRT-PCR) for the qualitative and quantitative detection of viral nucleic acids. Other relevant laboratory methods include enzyme-linked immunosorbent assays (ELISA) for viral antibody and antigen detection, and serum virus neutralization (SVN) assays for antibody neutralization determination. The challenges faced in developing a diagnostic test for a novel pathogen are the ability to measure low viral loads for early detection, to provide low or no cross-reactivity with other viral strains, and to deliver results rapidly. Several point-of-care molecular devices are currently being integrated for fast and accurate diagnosis of SARS-CoV-2 infections. This review discusses the current laboratory methods available to test for coronaviruses by focusing on the present COVID-19 outbreak.
Keywords: coronavirus, RT-PCR, ELISA, lateral flow diagnostics, convalescent plasma
Introduction
Coronavirus disease 2019 (COVID-19) is caused by a novel coronavirus (CoV) that was originally reported in Wuhan, Hubei province, China in December 2019.[1] The International Committee on Taxonomy of Viruses named the virus "severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Infection by SARS-CoV-2 causes a respiratory illness that varies in severity from mild upper respiratory symptoms akin to the seasonal flu, to severe progressive respiratory failure that requires intensive care and can lead to death. Asymptomatic carriers of the virus have also been reported and pose a significant public health threat due to their ability to unknowingly spread the virus.[2] SARS-CoV-2 represents the third coronavirus in this millennium to cross species from animals to humans and cause a severe respiratory disease, after Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012[3] and severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003.[4][5] SARS-CoV-2 has now been identified as the seventh coronavirus that is transmissible between humans (a group which also includes HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1).[6] On January 30, 2020, the World Health Organization (WHO) declared the SARS-CoV-2 epidemic a public health emergency of international concern, and the public health emergency was upgraded to a pandemic on March 11. At least 4,302,774 confirmed cases and 289,561 deaths worldwide were reported as of May 12.[7] Diagnostic testing is critical during a pandemic as the ability to track the spread of SARS-CoV-2 is essential for effective disease management and control.
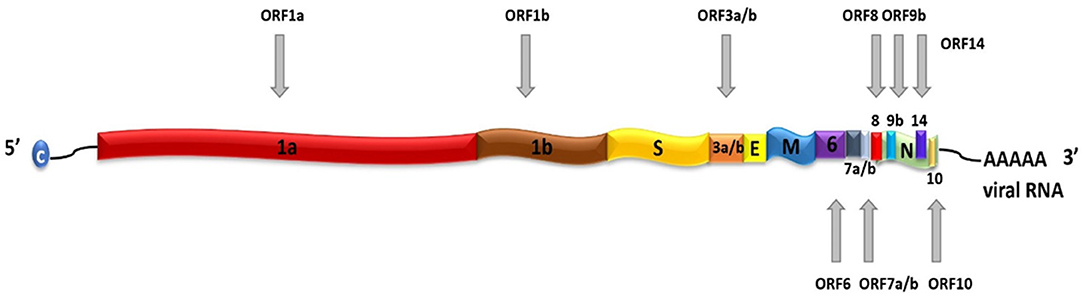
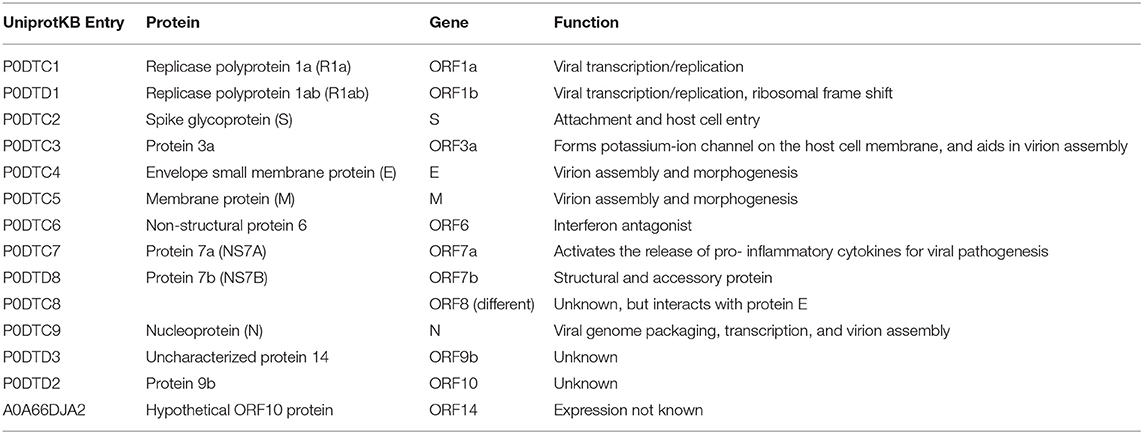
SARS-CoV-2 is a positive-sense, single-stranded RNA (ssRNA), group IV virus. The genome was sequenced from the bronchoalveolar lavage fluid of a patient (Genbank: MN908947) and shared through the Global Initiative on Sharing All Influenza Data (GISAID) platform on January 12, 2020.[8] The ~30 k base pair genome is highly similar to the human SARS-CoV and bat CoV-SARS-like genomes, with 14 open reading frames (ORFs) that encode structural, replication, and non-structural accessory proteins, as depicted in Figure 1. Molecular modeling studies demonstrate that like SARS-CoV, SARS-CoV-2 is surrounded by a lipid bilayer membrane, containing structural membrane (M) and envelope (E) proteins that interact to form the viral envelope.[9] This layer also contains spike glycoproteins (S) that give the characteristic “corona” appearance of this family of viruses. The spike proteins bind specific host cell receptors to facilitate host cell attachment and entry.[10] The nucleic acid-associated protein binds the RNA genome and forms the nucleocapsid (N). Other proteins include replication and non-structural accessory proteins, listed in Table 1. Reports of different strains of SARS-CoV-2 suggest an early split from the SARS-CoV-2 lineage and/or that the virus is mutating. Ongoing research provides insight into the unique and conserved features of the genome and proteome of SARS-CoV-2 for tracking mutations and generates evidence about the evolution of the virus.[11][12] This is important as these changes may affect key structural and non-structural components of SARS-CoV-2 that can render some diagnostic tests ineffective or less sensitive and can also impact the selection of epitopes for the development of new tests.
|
|
The spread of SARS-CoV-2 is primarily by respiratory droplets that arise from individuals that harbor the virus. Symptomatic individuals with the disease are one source of virus, but a major public health concern is transmission by mildly ill or asymptomatic individuals during the incubation period. Rapid viral diagnostic testing for SARS-CoV-2 is critical to identify these individuals and facilitate the implementation of protective measures such as social distancing, quarantine, and isolation that help to mitigate the spread of the virus in the community. The development of rapid and accurate tests that detect antibodies post-infection provide information about an individual's exposure to the virus and can be used to monitor the possibility of immunity, relapse, or reinfection. This supports interventions to protect high-risk populations from developing more severe illness and can be used to investigate the efficacy of passive antibody therapies for COVID-19 infection. This review describes the available testing methods for SARS-CoV-2 and brings to light the importance of laboratory testing to control this disease and prepare for possible future disease threats.
Detection of SARS-CoV-2 by real-time reverse transcription polymerase chain reaction (qRT-PCR)
The qRT-PCR method detects the genetic material of SARS-CoV-2 in order to identify the virus and/or quantify viral load. Comparisons of the ssRNA genetic sequences of this virus have shown similarities to SARS-CoV and several bat coronaviruses.[13] This detailed knowledge has allowed the rapid development of qRT-PCR assays for SARS-CoV-2 using SARS-CoV and other known coronaviruses as references.
Specifics
Sample collection
Upper and lower respiratory samples are collected. Nasopharyngeal swabs are high-priority specimens for SARS-CoV-2, and low-priority specimens include oropharyngeal swabs, bronchoalveolar lavage, tracheal aspirates, and sputum.[14]
RNA isolation
RNA is extracted from clinical specimens using approved viral isolation kits.[15]
qRT-PCR
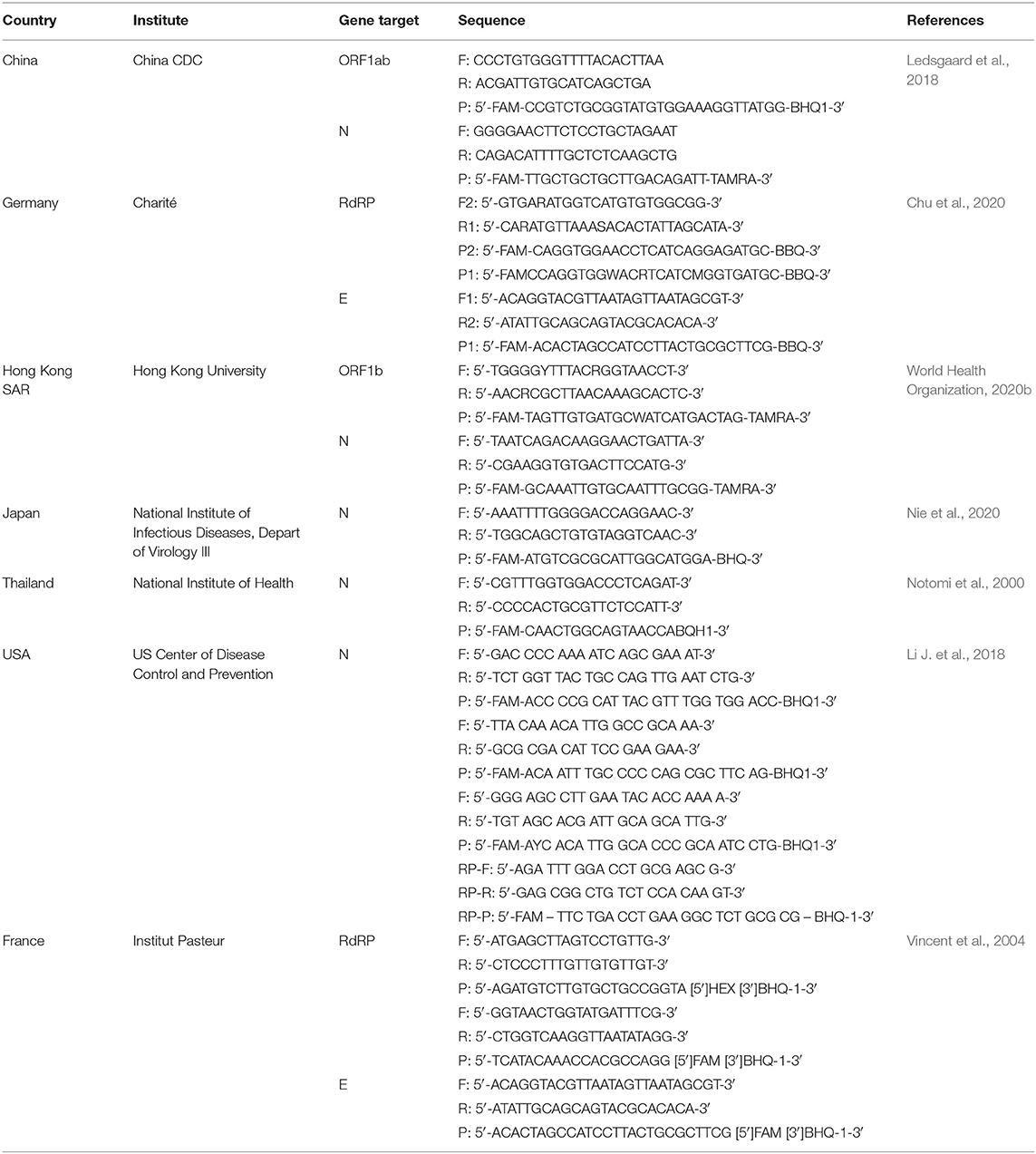
RNA is reverse transcribed to cDNA and subsequently amplified using a real-time quantitative PCR instrument. WHO announced various primer and probe sets for SARS-CoV-2 previously developed in China, Germany, Hong Kong, Japan, Thailand, and the United States (Table 2).[16] Primers target different sections of the virus' genetic sequence, including the envelope E gene, the RNA-dependent RNA polymerase (RdRp) gene, and the N gene.[15][16][17] Targeting the E gene is reported for highest sensitivity, followed by the RdRp gene for confirmation.[15] Some laboratories have multiplexed PCR tests consisting of multiple primer and probe sets located at different regions in the SARS-CoV-2 genome. These assays can be designed to contain primer sets targeting multiple genes simultaneously (RdRp/hel, S, N)[18], or to detect different regions in a single target such as the N gene (Table 2).[19] The use of multiplex assays is potentially beneficial as this can improve sensitivity in cases of loss or degradation of viral RNA during specimen collection and nucleic acid extraction, or in the event of mutation of the virus genome. These assays use in vitro synthesized RNA derived from transcripts (e.g., BetaCoV_Wuhan_WIV04_2019, GISAID Access number: EPI_ISL_402124) as positive controls and to generate standard curves. An internal control using RNAse P (RP) verifies the presence and quality of nucleic acid in samples, and molecular grade nuclease-free water is used as a negative amplification control. A negative patient sample serves both as a negative extraction control to monitor cross contamination across samples and to validate test reagents.
|
Advantages
qRT-PCR is the frontline diagnostic test for COVID-19, capable of analyzing thousands of specimens in a single day while showing a testing sensitivity of 95%.[15] The anticipated limit of detection of the SARS-CoV-2 qRT-PCR test is <10 copies/reaction[17], which allows early detection of low viral titers. Gene amplification indicates a positive result for the presence of SARS-CoV-2 RNA and should correlate with clinical observations, patient history, and epidemiological information.
Disadvantages
False positive results could be generated by cross-reactivity of primers with nucleic acids arising from co-infection with other viruses or bacteria. In these cases, the agent detected may not be the definite cause of disease. Matching of the SARS-CoV-2 qRT-PCR primers and probes using reliable libraries (e.g., BLAST) is necessary to ensure there is no homology with other coronaviruses like SARS-CoV from 2003 or other organisms such as Staphylococcus aureus and Candida albicans. False positives can also occur if reagents in a laboratory become contaminated, which is a major concern, particularly with the high volume of testing encountered during a pandemic. A negative patient sample is useful to identify this error in testing.
False negative results could potentially arise from mutations occurring in the primer and probe target regions in the SARS-CoV-2 genome. Negative results do not preclude SARS-CoV-2 infection, and results should be validated with different primer sets against the same gene target and combined with patient history and other clinical data to accurately determine patient infection status.
Key logistics
Provisions for testing laboratories, the use of approved tests, and validation of results with governing authorities to develop master protocols for use by multiple investigators must be in place to achieve rapid testing capacity. The output for number of tests per day and number of individuals tested per day relies on the availability of laboratory capacity, trained staff, reagents, supplies, and equipment. Large quantities of specific high-grade reagents are needed to perform tests, and supplies can be quickly depleted in a pandemic. This impacts the turnaround time for qRT-PCR diagnostic testing that ranges between two and five days. Strategies to rapidly scale up testing for novel human cornaviruses must be considered for future diagnostic testing.
Detection of antibodies against SARS-CoV-2 proteins by enzyme-linked immunosorbent assay (ELISA)
An ELISA is a diagnostic method used to identify antibodies in patient blood sample or nasopharyngeal swabs. An ELISA employed for antibody detection against SARS-CoV-2 measures the host humoral response, including IgM, IgG, and IgA to define previous exposure to the virus.[20][21] IgM is the first immunoglobulin that is produced in response to an antigen and is primarily detected during the early onset of disease (the first three to seven days). IgG is the most abundant immunoglobulin that is produced in response to an antigen (seven to 25 days) and is maintained in the body after initial exposure. It may also have a protective role for acquired immunity. The IgA immunoglobulin plays a crucial role in the immune function of mucous membranes.
The SARS-CoV-2 S glycoprotein that mediates attachment and entry into cells is surface-exposed and is a key target for the production of host-neutralizing antibodies.[22] This feature has made the S protein the focal target of antibody and vaccine development. The N protein in human coronaviruses' functions as an antagonist of interferon[23][24] and viral encoded repressor (VSR) of RNA interference (RNAi) that facilitates viral replication, and is also a key target for antibody design.[25] Recombinant antigens derived from the receptor binding domain of S protein (rS) as well as recombinant N protein (rN) are being developed as suitable diagnostic targets to detect IgM, IgG, and IgA antibodies. Dual detection of IgM/IgG and IgG/IgA immunoglobulins is under development for use in conjunction with nucleic acid detection for detecting active infection and to define previous exposure to SARS-CoV-2.
Specifics
Sample collection
Systemic blood samples are collected from individuals for extraction of serum.
ELISA
Purified rS or rN are immobilized to the surface of a multi-well plate as capture antigens. Controls and inactivated SARS-CoV-2 serum samples are incubated with the antigen for SARS-CoV-2 antibody-antigen binding. A labeled secondary antibody-conjugate (e.g., horseradish peroxidase) is bound to the SARS-CoV antibodies for signal detection by substrate addition, and quantification.
Advantages
Antibody tests provide the advantage of a simple method of detection of SARS-CoV-2 antibodies and are convenient to compare multiple samples from a single patient. Positive rates of detection for SARS-CoV-2 IgG in patients by ELISA measurements are 85.4% and 75.6–93.1% for IgM.[20] Jin et al.[26] reported sensitivities of serum IgM and IgG antibodies for detection were 48.1 and 88.9%, and specificities were 100 and 90.9%, with the highest sensitivity for antibody tests recorded two weeks after the first symptoms of disease. The lower IgM sensitivity may be because the IgM response occurs early then decreases and does not offer a strong detectable signal, while IgG signals may be more readily detected and present beyond 20 days. The incorporation of unique immunoglobulin labels may increase the sensitivity of rapid antibody tests for respiratory viruses.[27] Results from antibody testing could inform infection status and define previous exposure to SARS-CoV-2. Antibody detection is also used to identify recovered patients as human donors for the generation of convalescent patient serum or plasma as an investigational treatment for critically ill patients.[28]
Disadvantages
The results of SARS-CoV-2 antibody tests may vary by apparent disease periods and by time after symptom onset, as well as on the reliability of diagnostic assays. It is not yet known when IgM or IgG antibodies specific to the SARS-CoV-2 virus will become detectable during an infection, how long antibodies persist following infection, and what extent of protection neutralizing antibodies offer against subsequent infection with the virus.
The overall sensitivity and specificity indicate the possibility of false negatives and false positives in this testing method. Since the risk for recurrent infection with SARS-CoV-2 is not known for COVID-19, detection of one or two antibodies (IgM and/or IgG) does not necessarily guarantee immunity against reinfection. Negative results do not rule out SARS-CoV-2 infection—particularly in those who have been in contact with the virus—and positive results may be due to past or present infection with SARS-CoV[20] and possibly non-SARS-CoV strains.[29] It will be critical to conduct stringent evaluation of antibody diagnostic assays to determine the accuracy and reliability of results.
Key logistics
Recombinant systems are routinely used to express recombinant proteins to develop antibody assays. However, protein-expression systems can result in significant discrepancies between recombinant and native viral proteins. For example, the use of E. coli competent cells produces proteins that lack critical post-translational modifications in human cells (e.g., glycosylation) that can alter epitopes and protein conformation.[30] Consequently, this can compromise sensitivity and specificity of antigens for diagnostic assays. The use of mammalian expression systems to express recombinant proteins will produce antigens with post-translation modifications that more closely resemble human native proteins[31], leading to higher sensitivity and specificity of assays.
Serological assays are currently under accelerated development for diagnosis of human coronavirus infections. Commercial reagents need to be validated by clinical trials using samples from patients with confirmed infections of SARS-CoV-2, and approved by the regulatory review process. Nonetheless, a rapid and sensitive platform for identification of antibody titers will also support screening to identify and minimize the risk of viral spread to others, as well as for epidemiological studies and vaccine evaluation studies. The U.S. Food and Drug Administration (FDA) allows the use of rapid antibody tests for SARS-CoV-2 under emergency use authorization (EUA). This expedites the assessment and optimization of these diagnostic tests, with the expectation that any test is sufficiently experimentally validated before it is made available to patients. If these tests do not provide accurate results, this can impair prevention efforts and delay appropriate treatment during the global pandemic response.
Rapid detection of SARS-CoV-2 by lateral flow immunoassays (LFI)
Test developers have used the principles behind ELISA and applied them to develop lateral flow immunoassays (LFI) for the rapid qualitative detection of a substance. Some researchers are beginning to apply LFI to SARS-CoV-2 in the form of a simple, portable diagnostic strip to measure either SARS-CoV-2 antibodies or antigens. As viral titers are often low in nasal swabs and serum or plasma, detection of antigens may be more challenging in comparison to detection of antibodies. Serological antigen assays can target S1 and S2 domains of the S protein that binds angiotensin-converting enzyme-2 (ACE-2), an integral transmembrane protein in the lung alveolar epithelium that serves as the initial attachment site for SARS-CoV-2, or N proteins.
Specifics
LFI
The design of the lateral flow test is that of a strip/dipstick containing immobilized test reagents, enclosed in a cassette. Drops of a patient's blood are deposited on the strip, which contains a coating of purified monoclonal antibody (mAb) or recombinant antigen that is localized at specific regions on a nitrocellulose membrane. The mAb targets a viral antigen; the recombinant antigen is recognized by antibodies that are present in infected patients. The strip also contains labeled detector antibodies that bind the same antigen. A positive antibody result indicates binding between the coating antigen and patient antibodies and binding by the detector antibody. This generates a colored signal. A positive antigen result indicates binding between the coating antibody and patient antigen.
Advantage
Two drops of blood are sufficient for detection of SARS-CoV-2 and antibodies by this method. This technique delivers results in approximately 15 minutes and uses visual detection by the naked eye, in comparison to qRT-PCR (two to five days). Detection of antibodies shows previous viral exposure while detection of antigens indicates active carriers of SARS-CoV-2 virus. The specificity and sensitivity of LFIs are comparable with antibody and antigen assays.
Disadvantage
Tests to detect SARS-CoV-2 in patients by identifying viral antigens are more challenging to develop than tests to detect the neutralizing antibodies against SARS-CoV-2 (see below), as purified monoclonal antibodies must be generated against target antigens. Further, these assays need to be assessed and optimized using blood from infected patients.
Key logistics
The rapid development of some antigens for assays are led by the use of “prototype” pathogens and in silico models of antibody–antigen interactions that are used to generate artificial antibody libraries.[32] Antibody phage display technology can be applied to discover antibodies against antigens.[33] These can be rapidly generated to produce prototypes of diagnostic tests for validation studies that expedite assessment and optimization, before the final commercial diagnostic kits are available. Integrating fast, portable tests with epidemiological surveillance will also provide quick and reliable information to public health authorities monitoring the spread of SARS-CoV-2.
Serum virus neutralization assay (SVN)
The SVN assay is a serological test that measures the ability of a patient's antibodies to neutralize infectivity of SARS-CoV-2 and attenuate infection. This assay is considered the most reliable for the assessment of protective antibody and can inform the use of convalescent plasma as a passive antibody therapy for COVID-19 infection, particularly in severely ill patients. Although there is limited clinical data, early studies suggest that transfusion of convalescent plasma can suppress SARS-CoV-2 viral replication and protect an individual from infection.[20][28] The SVN assay is not used for routine diagnosis but is frontline for this special indication.
Specifics
Sample collection
Plasma is prepared from systemic blood samples collected from COVID-19 convalescent donors. Written informed consent is required from both the donor and recipient.
SVN
Several cell lines are suitable for SARS-CoV-2 transduction, including Vero (monkey kidney cell line), Huh7 (human hepatoma cell line), and 293T (human kidney cell line).[34] Serial dilutions of patient convalescent serum are added to known strains of virus (BetaCoV/Shenzhen/SZTH-003/2020 strain virus, GISAID access number: EPI_ISL_406594).[28] The mixture is inoculated into a susceptible cell monolayer and incubated for virus adsorption. The cytopathic effect can be measured by microscopic examination[28] after a five-day incubation, fluorescence[34], or plaque formation, following 24 hours of incubation. The neutralizing antibody titer is the highest dilution of serum that reduces activity of SARS-CoV-2.
Advantages
The SVN assay is a highly robust and reproducible test that may be applied to detect SARS-CoV-2 neutralizing antibodies in convalescent plasma samples to identify the best candidates for treatment. Neutralizing activities, along with viral load and antibody titers, can be simultaneously monitored in paired plasma samples in patients receiving convalescence plasma in order to establish algorithms for determining patient and donor factors that predict clinical efficacy.
Disadvantages
The accessibility of the live SARS-CoV-2 strain is regulated, which limits the development of laboratory testing by SVN. While inexpensive, it is a manual assay and requires careful in-house standardization and quality control.
Key logistics
A preliminary case report of positive responses of five severely ill patients with COVID-19 who were treated in the Shenzhen Third People's Hospital, China using plasma from recovered individuals was recently published.[28] The convalescent plasma contained functional IgG and IgM anti–SARS-CoV-2 neutralizing antibodies that inhibited viral growth in cell cultures. Notably, the SVN assay reliably measured the increases in the patients' neutralizing antibody titers between one and 12 days after plasma transfusion. This study was not evaluated in a randomized clinical trial, and there are limitations to the data interpretation. Nonetheless, these findings demonstrate the utility of the SVN assay for evaluating anti–SARS-CoV-2 neutralizing antibodies for future convalescent plasma assessment in more rigorous clinical investigations involving a larger cohort of patients with severe COVID-19 illness.
Emerging methods for diagnosis of SARS-CoV-2
Methods for the rapid detection of nucleic acids are being used to develop applications in clinical diagnostics of SAR-CoV-2.
Isothermal nucleic acid amplification
This method amplifies DNA or RNA target sequence in a streamlined and exponential manner for detection, and in contrast to PCR, does not require thermal cycling. A wide variety of nucleic acid detection assays have been developed, including loop-mediated isothermal amplification (LAMP), a single-tube technique for the amplification of DNA; reverse transcription-LAMP, which combines reverse transcriptase and LAMP to detect RNA (RT-LAMP)[35]; recombinase polymerase amplification (RPA)[36]; helicase-dependent amplification (HDA)[37]; strand displacement amplification (SDA)[38]; and nucleic acid sequence-based amplification (NASBA).[39] These assays incorporate isothermal methods to enable primer binding followed by amplification using a polymerase with strand-displacement activity that separates the strand that is annealed to the target sequence for detection. Amplified gene products can be detected by photometry. Isothermal nucleic acid amplification is utilized in several commercial molecular diagnostic platforms and is considered the fastest available molecular laboratory and point-of-care test for the detection of novel SARS-CoV-2.
RT-LAMP
The RT-LAMP method has been shown to effectively detect SARS-CoV-2 in clinical samples from individuals with COVID-19.[40] Multiple loop primers targeting the ORF1ab gene and the S gene were used for DNA strand displacement activity and target amplification that achieved detection of 20 copies/reaction and 200 copies/reaction, respectively. These results were comparable to qRT-PCR amplification. The reported 100% sensitivity and 100% specificity, as well as the mean time for detection being under 30 minutes, demonstrate this is a definitive testing method.
RPA
This method detected total viral RNA derived from cell culture supernatant and 19 nasopharyngeal swab samples (eight positive and 11 negative) for SARS-CoV-2.[41] This approach integrates isothermal methods for reverse transcription followed by recombinase activity that mediates primer (targeting the N gene) binding to the homologous sequence in dsDNA. Subsequent amplification by polymerase mediated primer extension achieved 100% diagnostic sensitivity and specificity. This method offers potential advantages over qRT-PCR for speed, scale, and portability, allowing evidence-based clinical decisions to be made during a patient visit.
CRISPR (clustered regularly interspaced short palindromic repeats)
The CRISPR assay functionality is being applied for detection of DNA or RNA using nucleic acid pre-amplification combined with CRISPR-Cas enzymology for specific recognition of sequences.
The CRISPR/Cas13a system is a recently discovered CRISPR-RNA (crRNA) guided detection method that is specific for RNA and is being applied for SARS-CoV-2 detection. A key feature of this approach is the Cas13a (formerly named C2c2) enzyme that recognizes and binds targeted RNA in a sequence-specific manner, followed by non-specific trans-endonuclease cleavage of non-targeted RNA (“collateral” cleavage) for signal amplification and nucleic acid detection. The Cas13a assay can be paired with target nucleic acid amplification for more sensitive results using an isothermal exponential amplification technique, most commonly RPA. This coupled technique is termed SHERLOCK (Specific High-Sensitivity Enzymatic Reporter unLOCKing) and allows fluorescence, colorimetric, lateral flow, and other readout approaches to enable the rapid detection of a variety of targets.[42]
Cas13a assay
Unlike in vivo CRISPR tools, the Cas13a protein must be recombinantly expressed and purified. The endonuclease activity of purified Cas13a uses crRNA targeting sequences in the S gene and ORF1ab in SARS-CoV-2 RNA. Target site-recognition activates trans-cleavage of reporter probes resulting in increases in fluorescence output signals and confirming the presence of viral RNA. Using synthetic SARS-CoV-2 the reported performance of this method for detection of target sequences is 20–200 aM.[42] The Cas13a/crRNA platform has been adapted for lateral-flow assays and could have wide applications as a SARS-CoV2 detector in both research and in the clinic. Assays can be designed as a paper dipstick test that delivers signals within 30 to 60 minutes. This is a very promising technology and these positive advances in science offer immense hope for future disease control.
Next-generation sequencing
Next-generation sequencing (NGS) enables complete sequencing of the ~30,000 nucleotides of the SARS-CoV-2 genome. NGS provides a method for identification of SARS-CoV-2, for environmental monitoring and surveillance testing, while also providing insight into strain origin and viral evolution. Each sequence is deposited into the GISAID EpiCoVTM Database, and to date there are over 17,000 SARS-CoV-2 sequences from global NGS efforts.
Sample is prepared by extracting RNA from clinical specimens, similar to qRT-PCR, and the sample is further purified to remove human cytoplasmic and ribosomal rRNA.
Library preparation involves fragmenting RNA, followed by cDNA synthesis. Through the use of a set of highly specific, universal coronavirus primers, all genomic segments are amplified and the DNA amplicons are sequenced to deliver highly accurate SARS-CoV-2 typing in less than 24 hours. Virus titer, efficiency of human rRNA depletion, and the number of reads per sample impact the number of virus-specific reads obtained and accurate coverage of the viral genome.
Collectively, global NGS data suggest the SARS-CoV-2 genome is relatively stable, although mutations are being identified in symptomatic individuals that are not present in the original strain in Wuhan, China. Two recent NGS studies report a large base pair deletion consisting of 81 nucleotides in SARS-CoV-2 ORF7a in a virus sample from a U.S. patient[43], and point mutations that may suggest a more infectious strain of the virus than the original strain.[44] The ORF7a gene encodes an accessory protein that is involved in viral infection and host cell death.[45] These findings require investigation in other patient samples and to determine whether such mutations are selected in asymptomatic or symptomatic individuals. Although NGS is one of the most comprehensive approaches for identifying SARS-CoV-2, this method is relatively expensive, with multiple sample preparation steps. As such, it tends to not be used for large-scale testing.
Biosafety
Regulating authorities provide interim guidance on the handling of specimens associated with SARS-CoV-2.[46] Samples for testing can be performed in a Biosafety Level 2 (BSL-2) laboratory with unidirectional airflow and BSL-3 precautions, and respiratory protection and a designated area for personal protective equipment changes are recommended. Isolation of SARS-CoV-2 in cell culture and initial characterization of viral agents recovered in cultures of patient samples should be conducted at Biosafety Level 3 (BSL-3), with regulatory approval and guidance.
Conclusions
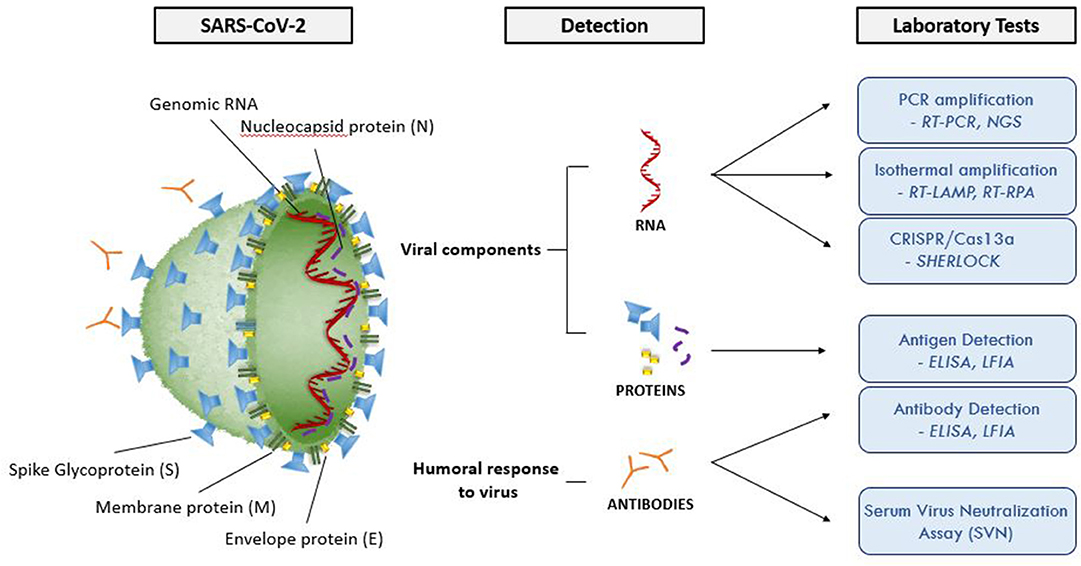
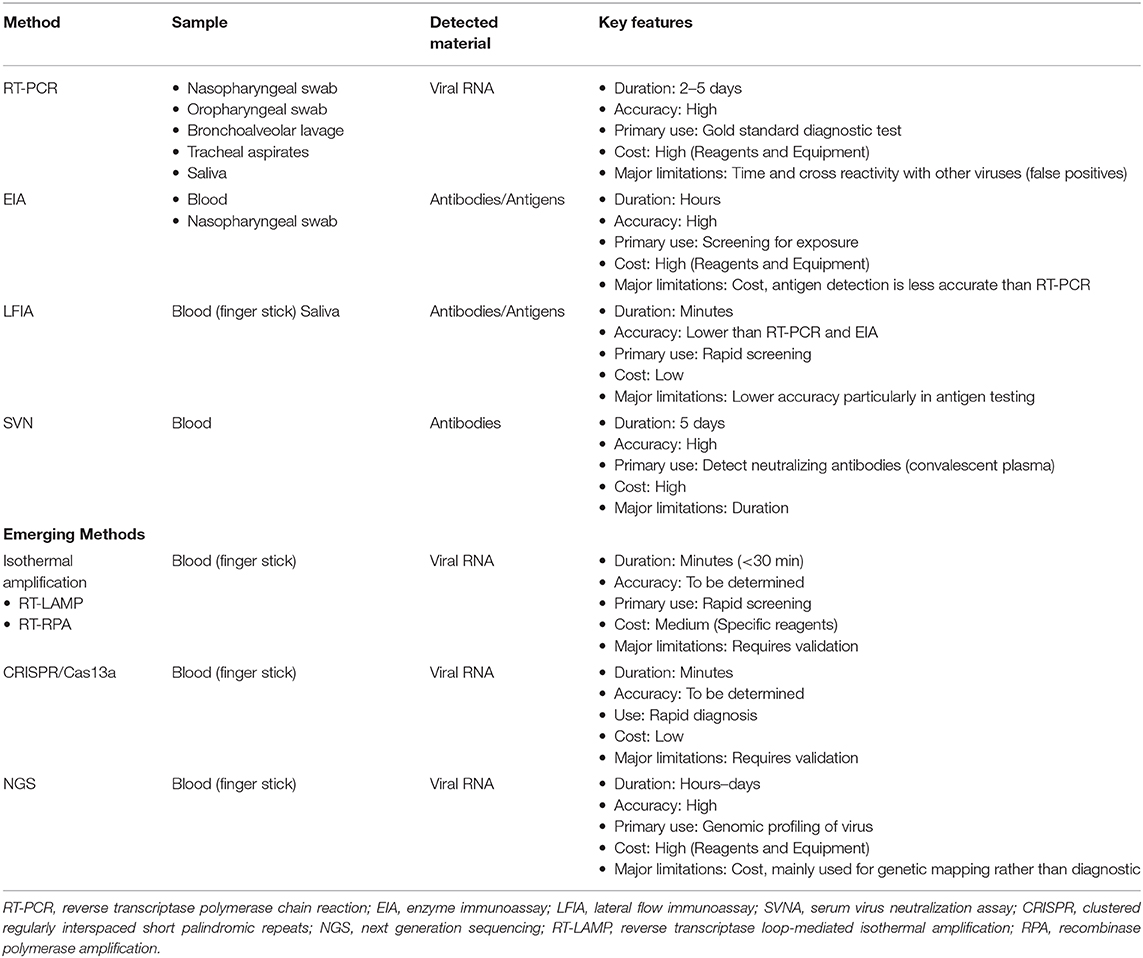
Figure 2 and Table 3 summarize the main laboratory tests for detection of components of SARS-CoV-2 and the humoral response to the virus, and they depict key features of these approaches. Given the public health emergency that the expanding COVID-19 outbreak presents, more widespread testing is needed to investigate the disease (e.g., prevalence in the population, severity in age groups) and to identify individuals who are infected but have few or no symptoms. Detailed epidemiological data sets will better establish the rates of severe infection and death among infected populations.
|
|
Ongoing research is critical to optimize existing antibody tests to determine whether immunity prevents recurrent infection and to investigate the efficacy of passive antibody therapies for COVID-19 infection. The identification of novel disease biomarkers may be valuable for understanding what makes people susceptible to COVID-19 infection and in predicting the severity and progression of disease. Researchers could request approval to analyze stored samples of human blood or in animals that might be a natural reservoir of the virus. Specifically, guidance would be needed to direct blood and plasma collection centers to allow access of samples from COVID-19 patients.
The COVID-19 pandemic showcases how quickly information needs to be shared as responders address rapidly evolving situations. Establishing communication across laboratories worldwide helps to develop master protocols and establish reference panels for use by multiple investigators. This aids in coordinating the collection and use of data within the constraints of the regulatory infrastructure. Having a range of tests also puts less pressure on one manufacturer or supply chain, as different suppliers may use different materials. This could help alleviate difficult decisions to limit testing to the most vulnerable patients, which can have great public health consequences.
Acknowledgements
The authors thank Robert E. Akins, Ph.D. (Nemours/A. I. duPont Hospital for Children) for reading and editing this manuscript.
Author contributions
RD'C and VS contributed to concept and writing of manuscript. AC contributed to writing, and prepared tables and figure for this manuscript.
Funding
This work was provided in part by Nemours/The Fund.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- ↑ World Health Organization (2020). "Naming the coronavirus disease (COVID-19) and the virus that causes it". World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it. Retrieved May 2020.
- ↑ Chan, J.F.-W.; Yuan, S.; Kok, K.-H. et al. (2020). "A Familial Cluster of Pneumonia Associated With the 2019 Novel Coronavirus Indicating Person-To-Person Transmission: A Study of a Family Cluster". Lancet 395 (10223): 514–23. doi:10.1016/S0140-6736(20)30154-9. PMC PMC7159286. PMID 31986261. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7159286.
- ↑ Zaki, A.M.; van Boheemen, S.; Besterbroer, T.M. et al. (2012). "Isolation of a Novel Coronavirus From a Man With Pneumonia in Saudi Arabia". New England Journal of Medicine 367 (19): 1814–20. doi:10.1056/NEJMoa1211721. PMID 23075143.
- ↑ Drosten, C.; Günther, S.; Preiser, W. et al. (2003). "Identification of a Novel Coronavirus in Patients With Severe Acute Respiratory Syndrome". New England Journal of Medicine 348 (20): 1967-76. doi:10.1056/NEJMoa030747. PMID 12690091.
- ↑ Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S. et al. (2003). "A Novel Coronavirus Associated With Severe Acute Respiratory Syndrome". New England Journal of Medicine 348 (20): 1953–66. doi:10.1056/NEJMoa030781. PMID 12690092.
- ↑ Salata, C.; Calistri, A.; Parolin, C. et al. (2019). "Coronaviruses: A Paradigm of New Emerging Zoonotic Diseases". Pathogens and Disease 77 (9): ftaa006. doi:10.1093/femspd/ftaa006. PMC PMC7108526. PMID 32065221. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7108526.
- ↑ "COVID-19 Coronavirus Pandemic". Worldometer. https://www.worldometers.info/coronavirus/. Retrieved 12 May 2020. >
- ↑ Wu, F.; Zhao, S.; Yu, B. et al. (2020). "A New Coronavirus Associated With Human Respiratory Disease in China". Nature 579 (7798): 265-269. doi:10.1038/s41586-020-2008-3. PMC PMC7094943. PMID 32015508. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7094943.
- ↑ Durrant, J.D.; Kochanek, S.E.; Casalino, L. et al. (2020). "Mesoscale All-Atom Influenza Virus Simulations Suggest New Substrate Binding Mechanism". ACS Central Science 6 (2): 189–96. doi:10.1021/acscentsci.9b01071. PMC PMC7048371. PMID 32123736. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7048371.
- ↑ Graham, R.L.; Baric, R.S. (2010). "Recombination, Reservoirs, and the Modular Spike: Mechanisms of Coronavirus Cross-Species Transmission". Journal of Virology 84 (7): 3134–46. doi:10.1128/JVI.01394-09. PMC PMC2838128. PMID 19906932. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2838128.
- ↑ Phan, T. (2020). "Genetic Diversity and Evolution of SARS-CoV-2". Infection, genetics and evolution 81: 104260. doi:10.1016/j.meegid.2020.104260. PMC PMC7106203. PMID 32092483. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7106203.
- ↑ Wang, C.; Liu, Z.; Chen, Z. et al. (2020). "The Establishment of Reference Sequence for SARS-CoV-2 and Variation Analysis". Journal of Medical Virology 92 (6): 667-674. doi:10.1002/jmv.25762. PMC PMC7228400. PMID 32167180. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7228400.
- ↑ Lu, R.; Zhao, X.; Li, J. et al. (2020). "Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding". Lancet 395 (10224): 565–74. doi:10.1016/S0140-6736(20)30251-8. PMC PMC7159086. PMID 32007145. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7159086.
- ↑ Centers for Disease Control and Prevention (22 May 2020). "Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens for COVID-19". Centers for Disease Control and Prevention. https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html. Retrieved May 2020.
- ↑ 15.0 15.1 15.2 15.3 Corman, V.M.; Landt, O.; Kaiser, M. et al. (2020). "Detection of 2019 Novel Coronavirus (2019-nCoV) by Real-Time RT-PCR". Euro Surveillance 25 (3): 2000045. doi:10.2807/1560-7917.ES.2020.25.3.2000045. PMC PMC6988269. PMID 31992387. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6988269.
- ↑ 16.0 16.1 World Health Organization (2020). "Coronavirus disease (COVID-19) technical guidance: Laboratory testing for 2019-nCoV in humans". World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance. Retrieved May 2020.
- ↑ 17.0 17.1 Chu, D.K.W.; Pan, Y.; Cheng, S.M.S. et al. (2020). "Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia". Clinical Chemistry 66 (4): 549-555. doi:10.1093/clinchem/hvaa029. PMC PMC7108203. PMID 32031583. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7108203.
- ↑ Chan, J.F.-K.; Yip, C.C.-Y.; To, K.K.-W. et al. (2020). "Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and With Clinical Specimens". Journal of Clinical Microbiology 58 (5): e00310-20. doi:10.1128/JCM.00310-20. PMC PMC7180250. PMID 32132196. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7180250.
- ↑ U.S. Food and Drug Administration (2020). "Orig3n 2019 Novel Coronavirus (COVID-19) Test EUA Summary". U.S. Food and Drug Administration. https://www.fda.gov/media/136873/download. Retrieved May 2020.
- ↑ 20.0 20.1 20.2 20.3 Guo, L.; Ren, L.; Yang, S. et al. (2020). "Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19)". Clinical Infectious Diseases: ciaa310. doi:10.1093/cid/ciaa310. PMC PMC7184472. PMID 32198501. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7184472.
- ↑ Okba, N.M.A.; Müller, M.A.; Li, W. et al. (2020). "Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease 2019 Patients". Emerging Infectious Diseases 26 (7). doi:10.3201/eid2607.200841. PMID 32267220.
- ↑ Walls, A.C.; Tortorici, M.A.; Frenz, B. et al. (2016). "Glycan Shield and Epitope Masking of a Coronavirus Spike Protein Observed by Cryo-Electron Microscopy". Nature Structural and Molecular Biology 23 (10): 899-905. doi:10.1038/nsmb.3293. PMC PMC5515730. PMID 27617430. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5515730.
- ↑ Kopecky-Bromberg, S.A.; Martínez-Sobrido, L.; Frieman, M. et al. (2007). "Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists". Journal of Virology 81 (2): 548-57. doi:10.1128/JVI.01782-06. PMC PMC1797484. PMID 17108024. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1797484.
- ↑ McBride, R.; van Zyl, M.; Fielding, B.C. et al. (2014). "The Coronavirus Nucleocapsid Is a Multifunctional Protein". Viruses 6 (8): 2991-3018. doi:10.3390/v6082991. PMC PMC4147684. PMID 25105276. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4147684.
- ↑ Leung, D.T.M.; Tam, F.C.H.; Ma, C.H. et al. (2004). "Antibody Response of Patients With Severe Acute Respiratory Syndrome (SARS) Targets the Viral Nucleocapsid". Journal of Infectious Diseases 190 (2): 379-86. doi:10.1086/422040. PMC PMC7110057. PMID 15216476. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7110057.
- ↑ Jin, Y.; Wang, M.; Zuo, Z. et al. (2020). "Diagnostic Value and Dynamic Variance of Serum Antibody in Coronavirus Disease 2019". International Journal of Infectious Diseases 94: 49–52. doi:10.1016/j.ijid.2020.03.065. PMC PMC7194885. PMID 32251798. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7194885.
- ↑ Li, R.; Tian, X.; Yu, Y. et al. (2018). "Development of a Rapid Immunochromatographic Assay for Detection of Antibodies Against Porcine Epidemic Diarrhea Virus". Polish Journal of Veterinary Sciences 21 (1): 139-147. doi:10.24425/119032. PMID 29624018.
- ↑ 28.0 28.1 28.2 28.3 28.4 Shen, C.; Wang, Z.; Zhao, F. et al. (2020). "Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma". JAMA 323 (16): 1582-1589. doi:10.1001/jama.2020.4783. PMC PMC7101507. PMID 32219428. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7101507.
- ↑ Gaunt, E.R.; Hardie, A.; Claas, E.C.J. et al. (2010). "Epidemiology and Clinical Presentations of the Four Human Coronaviruses 229E, HKU1, NL63, and OC43 Detected Over 3 Years Using a Novel Multiplex Real-Time PCR Method". Journal of Clinical Microbiology 48 (8): 2940-7. doi:10.1128/JCM.00636-10. PMC PMC2916580. PMID 20554810. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2916580.
- ↑ Gupta, S.J.; Shukla, P. (2018). "Glycosylation Control Technologies for Recombinant Therapeutic Proteins". Applied Microbiology and Biotechnology 102 (24): 10457-10468. doi:10.1007/s00253-018-9430-6. PMID 30334089.
- ↑ Bandaranayak, A.D.; Almo, S.C. (2014). "Recent Advances in Mammalian Protein Production". FEBS Letters 588 (2): 253-60. doi:10.1016/j.febslet.2013.11.035. PMID PMC3924552.
- ↑ Shao, Y.-M.; Yang, W.-B.; Peng, H.-P. et al. (2007). "Structure-based Design and Synthesis of Highly Potent SARS-CoV 3CL Protease Inhibitors". Chembiochem 8 (14): 1654-7. doi:10.1002/cbic.200700254. PMID PMC7162026.
- ↑ Ledsgarrd, L.; Kilstrup, M.; Karatt-Vellatt, A. et al. (2018). "Basics of Antibody Phage Display Technology". Toxins 10 (6): 236. doi:10.3390/toxins10060236. PMID PMC6024766.
- ↑ 34.0 34.1 Nie, J.; Li, Q.; Wu, J. et al. (2020). "Establishment and Validation of a Pseudovirus Neutralization Assay for SARS-CoV-2". Emerging Microbes & Infections 9 (1): 680–86. doi:10.1080/22221751.2020.1743767. PMID PMC7144318.
- ↑ Notomi, T.; Okayama, H.; Masubuchi, H. et al. (2020). "Loop-mediated Isothermal Amplification of DNA". Nucleic Acids Research 28 (12): E63. doi:10.1093/nar/28.12.e63. PMID PMC102748.
- ↑ Li, J.; Macdonald, J.; von Stetten, F. (2018). "Review: A Comprehensive Summary of a Decade Development of the Recombinase Polymerase Amplification". The Analyst 144 (1): 31–67. doi:10.1039/c8an01621f. PMID 30426974.
- ↑ Vincent, M.; Xu, Y.; Kong, H. (2004). "Helicase-dependent Isothermal DNA Amplification". EMBO Reports 5 (8): 795–800. doi:10.1038/sj.embor.7400200. PMC PMC1249482. PMID 15247927. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1249482.
- ↑ Walker, G.T.; Fraiser, M.S.; Schram, J.L. et al. (1992). "Strand Displacement Amplification--An Isothermal, in Vitro DNA Amplification Technique". Nucleic Acids Research 20 (7): 1691-6. doi:10.1093/nar/20.7.1691. PMC PMC312258. PMID 1579461. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC312258.
- ↑ Compton, J. (1991). "Nucleic Acid Sequence-Based Amplification". Nature 350 (6313): 91-2. doi:10.1038/350091a0. PMID 1706072.
- ↑ Yan, C.; Cui, J.; Huang, L. et al. (2020). "Rapid and Visual Detection of 2019 Novel Coronavirus (SARS-CoV-2) by a Reverse Transcription Loop-Mediated Isothermal Amplification Assay". Clinical Microbiology and Infection 26 (6): 773–79. doi:10.1016/j.cmi.2020.04.001. PMC PMC7144850. PMID 32276116. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7144850.
- ↑ Behrmann, O.; Bchmann, I.; Spiegel, M. et al. (2020). "Rapid Detection of SARS-CoV-2 by Low Volume Real-Time Single Tube Reverse Transcription Recombinase Polymerase Amplification Using an Exo Probe With an Internally Linked Quencher (exo-IQ)". Clinical Chemistry: hvaa116. doi:10.1093/clinchem/hvaa116. PMC PMC7239256. PMID 32384153. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7239256.
- ↑ 42.0 42.1 Kellner, M.J.; Koob, J.G.; Gootenberg, J.S. et al. (2019). "SHERLOCK: Nucleic Acid Detection With CRISPR Nucleases". Nature Protocols 14 (10): 2986–3012. doi:10.1038/s41596-019-0210-2. PMC PMC6956564. PMID 31548639. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6956564.
- ↑ Holland, L.A.; Kaelin, E.A.; Maqsood, R. et al. (2020). "An 81 Nucleotide Deletion in SARS-CoV-2 ORF7a Identified From Sentinel Surveillance in Arizona (Jan-Mar 2020)". Journal of Virology: JVI.00711-20. doi:10.1128/JVI.00711-20. PMID 32357959.
- ↑ Korber, B.; Fischer, W.M.; Gnanakaran, S. et al. (2020). "Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2". bioRxiv. doi:10.1101/2020.04.29.069054.
- ↑ Schaecher, S.R.; Touchette, E.; Schriewer, J. et al. (2007). "Severe Acute Respiratory Syndrome Coronavirus Gene 7 Products Contribute to Virus-Induced Apoptosis". Journal of Virology 81 (20): 11054-68. doi:10.1128/JVI.01266-07. PMC PMC2045523. PMID 17686858. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2045523.
- ↑ Centers for Disease Control and Prevention (May 2020). "Interim Laboratory Biosafety Guidelines for Handling and Processing Specimens Associated with Coronavirus Disease 2019 (COVID-19)". Centers for Disease Control and Prevention. https://www.cdc.gov/coronavirus/2019-ncov/lab/lab-biosafety-guidelines.html. Retrieved May 2020.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. References in this version are listed in order of appearance—by design—rather than alphabetical order as the original was.