Journal:Why health services research needs geoinformatics: Rationale and case example
| Full article title | Why health services research needs geoinformatics: Rationale and case example |
|---|---|
| Journal | Journal of Health & Medical Informatics |
| Author(s) | Onega, Tracy; Alford-Teaster, Jennifer; Andrews, Steven; Ganoe, Craig; Perez, Mike; King, David; Shi, Xun |
| Author affiliation(s) | Geisel School of Medicine at Dartmouth; Exaptive, Inc.; Dartmouth College |
| Primary contact | Email: jennifer.a.alford-teaster@dartmouth.edu; Phone: 603-653-6088 |
| Year published | 2014 |
| Volume and issue | 5 (6) |
| Page(s) | 176 |
| DOI | 10.4172/2157-7420.1000176 |
| ISSN | 2157-7420 |
| Distribution license | Creative Commons Attribution License (journal fails to specify which version) |
| Website | http://www.omicsonline.org/open-access/ |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Introduction
Delivery of health care in the United States has become increasingly complex over the past 50 years, as health care markets have evolved, technology has diffused, population demographics have shifted, and cultural expectations of health and health care have been transformed. Identifying and understanding important patterns of health care services, accessibility, utilization, and outcomes can best be accomplished by combining data from all of these dimensions in near-real time. The Big Data paradigm provides a new framework to bring together very large volumes of data from a variety of sources and formats, with computing capacity to derive new information, hypotheses, and inferences.[1][2] The complementary fields of genomics and bioinformatics have already made great advances only made possible by Big Data approaches. Similar gains can be made by pairing health services research with geoinformatics –- defined as “the science and technology dealing with the structure and character of spatial information, its capture, its classification and qualification, its storage, processing, portrayal and dissemination, including the infrastructure necessary to secure optimal use of this information”.[3] Integrating geospatial technologies with health services research brings informatics approaches, data sciences, and spatial theories of health and healthcare together to explore relationships among geography, health, and delivery of care in novel ways made possible through geoinformatics. Syngergy between the two disciplines will enhance our ability to discover how health care is delivered most effectively for the greatest health benefits across populations.
Health services research and geography have intersected under the rubrics of medical geography, epidemiology of health care, small area analysis, and public health since ancient Roman times, but more formally since the 1930s. Then, James Glover — an English physician — noticed that tonsillectomies were occurring at highly variable rates across school districts, which could not be explained by geographic, socio-demographic, or clinical factors, and the most likely explanation suggested being differences in how physicians practice.[4] Similar work, begun in the 1970s, became the impetus for the Dartmouth Atlas of Health Care[5], which used health care utilization data to develop geographic units representing health care markets. These health care-based spatial units can be directly compared to identify patterns associated with both effective and ineffective care. Other notable examples of health services research linked to a geospatial framework can be found in the public health arena, with planning of population-based vaccination programs and designation of federally-qualified health centers. Despite these tremendous contributions, limitations exist that geoinformatics and health service research are now poised to overcome as information technology and the digital era expands data availability, accessibility, usability, and timely knowledge generation.
Understanding spatiotemporal distributions of health services is a fundamental aspect of health services research upon which studies of utilization, outcomes, comparative effectiveness, resource allocation, and others are based. Four key limitations can be found in typical approaches to measuring health services distributions: 1. retrospective methods; 2. limited geographic extents; 3. ascertainment challenges; and 4. structured data only; which will be addressed below (Table 1). By combining geoinformatics with health services research, an important domain of questions within the field of medical informatics can be addressed, such as is illustrated with a case example.
|
- Table 1: Summary of major limitations in geographical based health services research and how geoinformatics approaches can be used to address them
A health services research problem: Diffusion of medical technology
Technological innovation is a hallmark of the U.S. health care system, and relies on the backbone of translational research to evaluate effectiveness after efficacy has been established. The full potential of a new technology is determined as clinical improvements and impacts on population health are assessed. Yet typically diffusion and the research to establish effectiveness occur asynchronously. This limits the potential for timely assessment of broad clinical impact and leads to two concerns: 1. overuse of technologies with unproven or minimal benefits in the general population and concomitant unwarranted costs; 2. underuse of technologies that have beneficial effects and improve outcomes for particular patient populations. As medical technology diffuses into generalizable practice, data and research needs arise to address these concerns from a host of stakeholders perspectives, including patients, clinicians, researchers, health care facilities, commercial vendors, payers, health care systems, communities, and regulatory bodies. A prerequisite for assessing new technologies in community practice is knowledge of the locations or extent of diffusion and the populations reached. Data availability and timeliness are critical barriers to establishing this knowledge, contributing to spotty information that is retrospective, often with notable lags. For example, The Dartmouth Atlas of Healthcare[5] and other work[6][7][8] has described geographic variation of medical technologies at a national level, but relied on Medicare data, which are typically available for research with a lag of 2-3 years. Further, Medicare, as well as private insurers, are dependent on the new technology being approved for reimbursement since they rely on billing data (claims), and coverage of the technology may take years following FDA approval and commercial dissemination, thus not able to be ascertained until unique billing codes are implemented. Some data resources may be timely for monitoring diffusion — such as registries like the Breast Cancer Surveillance Consortium (BCSC) and the HMO Research Network (HMORN)[9][10], and other clinical/provider networks with health information exchanges and/or robust electronic health records. However, these data resources are limited in geographic extent, are often not population-based, and may not capture use of new technologies until they are uniquely coded (e.g. CPT – Common Procedural Terminology), or are reimbursable. Further, most data sources require existing structured data, although natural language processing (NLP) is increasingly applied to unstructured medical record information, which can be powerful, but also time-consuming, complicated and variable across settings.[11][12][13] Achieving national extent, fully ascertained, and timely data capture to characterize geographic and sociodemographic diffusion of new technologies is a critical need, particularly as the U.S. seeks to improve health and healthcare and limit health care costs to effective use within populations.
Measuring near-real time diffusion of breast imaging technology
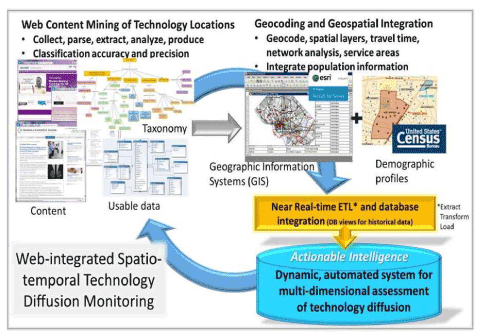
To more fully understand dissemination of new technologies we need to be able to capture the occurrences of the technology in large geographic areas, and in a dynamic way that reflects the dynamic process that dissemination is. Figure 1 presents a schematic approach to do this, using a breast imaging technology diffusion cased example (Figure 1). We can address the existing limitations in measuring, monitoring, and characterizing health care diffusion — with breast imaging as a timely example, given new technologies, such as digital breast tomosynthesis (DBT)[14][15][16], and legislation related to breast density notification [17]. With a geospatial semantic web [18-22], which combines web mining techniques with geographic information systems and census data, one can ascertain geographic uptake of DBT nationally, estimate potential access overall and by population subgroups, and identify correlates of dissemination patterns.
Figure 1. Schematic flow for a geoinformatics application in health services<br /research to dynamically monitor technology diffusion
Web content mining is used in this project to identify instances of DBT based on taxonomy of terms. Using associated web pages from these instances, street address information is captured for the DBT instance. These addresses and related attributes (facility name, date of data capture, etc.) are brought into a GIS for geocoding, spatial joins with other layers (e.g. road network), travel time analysis, and service area creation. Population demographics and other census-derived data are attributed to DBT locations and service areas. This constitutes the database containing geographic extent of DBT diffusion and population characteristics served by DBT facilities, which is refreshed at regular intervals following external validation. This application characterizes heterogeneous diffusion both geographically and sociodemographically, which also serves as a proof of concept for higher-dimension data which incorporates spatial layers, as well as other technologies.
Conclusion
Acknowledgements
References
- ↑ Philip Chen, C.L.; Zhang, C.Y. (2014). "Data-intensive applications, challenges, techniques and technologies: A survey on Big Data". Information Sciences 275: 314-347. doi:10.1016/j.ins.2014.01.015.
- ↑ Kambatla, K.; Kolliasb, G.; Kumar, V.; Grama, A. (2014). "Trends in big data analytics". Journal of Parallel and Distributed Computing 74 (7): 2561-2573. doi:10.1016/j.jpdc.2014.01.003.
- ↑ Raju, P.L.N. (2004). "Fundamentals of Geographic Information Systems" (PDF). Satellite Remote Sensing and GIS Applications in Agricultural Meteorology. World Meteorological Organisation. http://www.preventionweb.net/files/1682_9970.pdf.
- ↑ Glover, J.A. (1938). "The incidence of tonsillectomy in school children". Proceedings of the Royal Society of Medicine 31 (10): 95-113. PMC PMC2076749. PMID 18245048. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2076749.
- ↑ 5.0 5.1 "The Dartmouth Atlas of Health Care". The Dartmouth Institute. http://www.dartmouthatlas.org/publications/articles.aspx.
- ↑ Onega, T.; Toseston, T.D.; Wang, Q.; Hillner, B.E.; Song, Y.; Siegel, B.A.; Tosteson, A.N. (2012). "Geographic and sociodemographic variation of PET use in Medicare beneficiaries with cancer". Journal of the American College of Radiology 9 (9): 635-642. doi:10.1016/j.jacr.2012.05.005. PMC PMC3830950. PMID 22954545. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3830950.
- ↑ Onega, T.; Hubbard, R.; Hill, D.; Lee, C.I.; Haas, J.S.; Carlos, H.A.; Alford-Teaster, J.; Bogart, A.; DeMartini, W.B.; Kerlikowske, K.; Virnig, B.A.; Buist, D.S.; Henderson, L.; Tosteson, A.N. (2014). "Geographic Access to Breast Imaging for U.S. Women". Journal of the American College of Radiology 11 (9): 874-882. doi:10.1016/j.jacr.2014.03.022. PMC PMC4156905. PMID 24889479. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4156905.
- ↑ Onega, T.; Duell, E.J.; Shi, X.; Wang, D.; Demidenko, E.; Goodman, D. (2008). "Geographic access to cancer care in the U.S.". Cancer 112 (4): 909-918. doi:10.1002/cncr.23229. PMID 18189295.
- ↑ "Breast Cancer Surveillance Consortium". National Cancer Institute. http://breastscreening.cancer.gov/about/.
- ↑ "HMO Research Network". Kaiser Permanente Division of Research. http://www.hcsrn.org/en/About/.
- ↑ Xu, R.; Garten, Y.; Supekar, K.S.; Das, A.K.; Altman, R.B.; Garber, A.M. (2007). "Extracting subject demographic information from abstracts of randomized clinical trial reports". MEDINFO 2007 129: 550-554. PMID 17911777.
- ↑ Xu, R.; Superkar, K.; Huang, Y.; Das, A.K.; Garber, A.M. (2006). "Combining text classification and Hidden Markov Modeling techniques for structuring randomized clinical trial abstracts". AMIA Annual Symposium Proceedings 2006: 824-828. PMC PMC1839538. PMID 17238456. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1839538.
- ↑ Embley, D.W. (2004). "Toward semantic understanding: An approach based on information extraction ontologies". Proceedings of the Fifteenth Australasian Database Conference 27: 3-12.
- ↑ Dobbins, J.T. (2009). "Tomosynthesis imaging: at a translational crossroads". Medical Physics 36 (6): 1956-1967. PMC PMC2832060. PMID 19610284. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2832060.
- ↑ Kopans, D.B. (2014). "Digital breast tomosynthesis from concept to clinical care". American Journal of Roentgenology 202 (2): 299-308. doi:10.2214/AJR.13.11520. PMID 24450669.
- ↑ Lee, C.I.; Lehman, C.D. (2013). "Digital breast tomosynthesis and the challenges of implementing an emerging breast cancer screening technology into clinical practice". Journal of the American College of Radiology 10 (12): 913-917. doi:10.1016/j.jacr.2013.09.010. PMID 24295940.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In most of the article's references DOIs and PubMed IDs were not given; they've been added to make the references more useful.