Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Panahiazar JofBiomedInformatics2017 72-8.jpg|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Predicting biomedical metadata in CEDAR: A study of Gene Expression Omnibus (GEO)|Predicting biomedical metadata in CEDAR: A study of Gene Expression Omnibus (GEO)]]"''' | ||
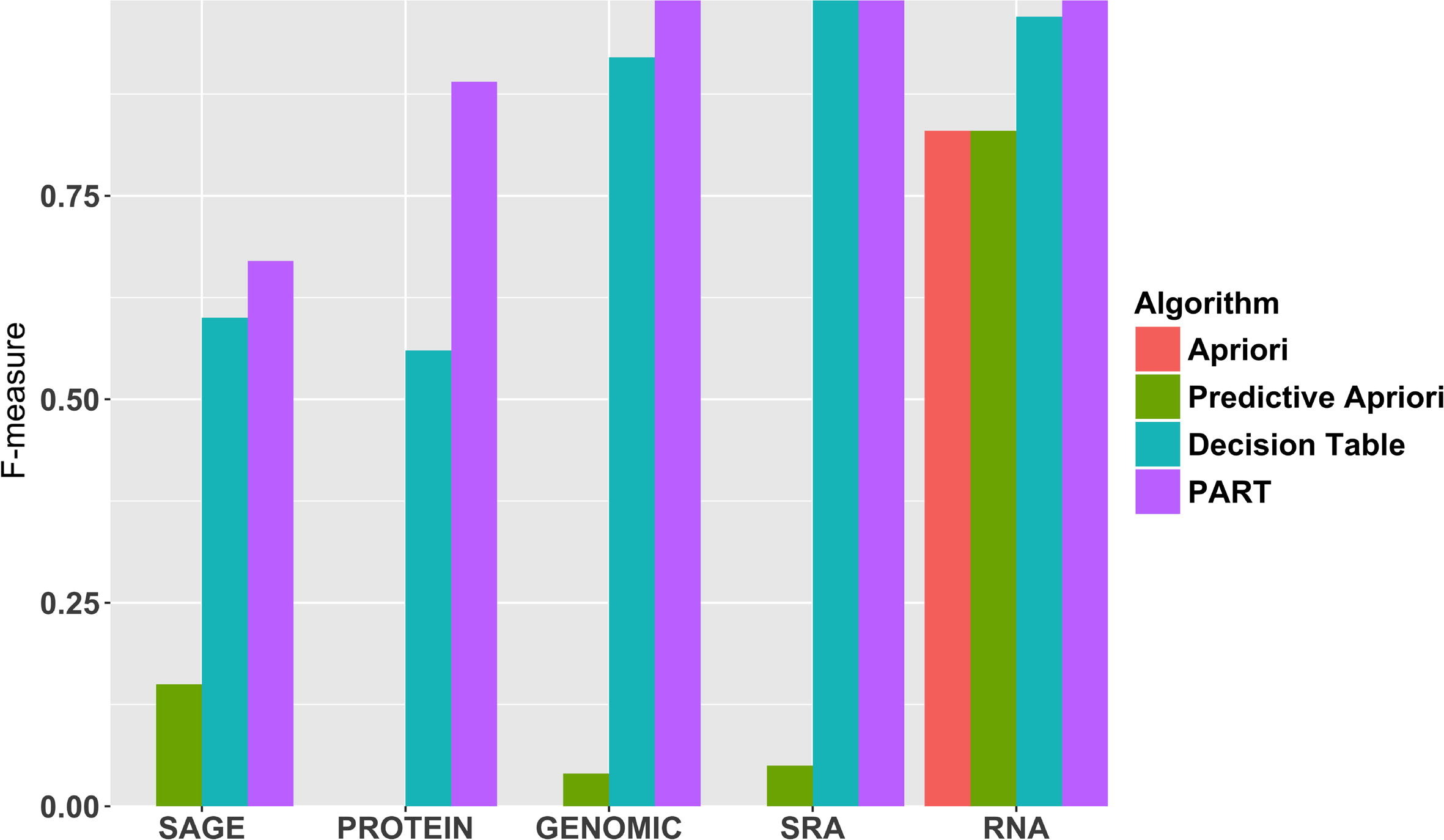
A crucial and limiting factor in data reuse is the lack of accurate, structured, and complete descriptions of data, known as metadata. Towards improving the quantity and quality of metadata, we propose a novel metadata prediction framework to learn associations from existing metadata that can be used to predict metadata values. We evaluate our framework in the context of experimental metadata from the Gene Expression Omnibus (GEO). We applied four rule mining algorithms to the most common structured metadata elements (sample type, molecular type, platform, label type and organism) from over 1.3 million GEO records. We examined the quality of well supported rules from each algorithm and visualized the dependencies among metadata elements. Finally, we evaluated the performance of the algorithms in terms of accuracy, precision, recall, and F-measure. We found that PART is the best algorithm outperforming Apriori, Predictive Apriori, and Decision Table. | |||
All algorithms perform significantly better in predicting class values than the majority vote classifier. We found that the performance of the algorithms is related to the dimensionality of the GEO elements. ('''[[Journal:Predicting biomedical metadata in CEDAR: A study of Gene Expression Omnibus (GEO)|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Rapid development of entity-based data models for bioinformatics with persistence object-oriented design and structured interfaces|Rapid development of entity-based data models for bioinformatics with persistence object-oriented design and structured interfaces]] | |||
: ▪ [[Journal:Bioinformatics education in pathology training: Current scope and future direction|Bioinformatics education in pathology training: Current scope and future direction]] | : ▪ [[Journal:Bioinformatics education in pathology training: Current scope and future direction|Bioinformatics education in pathology training: Current scope and future direction]] | ||
: ▪ [[Journal:FluxCTTX: A LIMS-based tool for management and analysis of cytotoxicity assays data|FluxCTTX: A LIMS-based tool for management and analysis of cytotoxicity assays data]] | : ▪ [[Journal:FluxCTTX: A LIMS-based tool for management and analysis of cytotoxicity assays data|FluxCTTX: A LIMS-based tool for management and analysis of cytotoxicity assays data]] | ||
Revision as of 17:00, 17 October 2017
"Predicting biomedical metadata in CEDAR: A study of Gene Expression Omnibus (GEO)"
A crucial and limiting factor in data reuse is the lack of accurate, structured, and complete descriptions of data, known as metadata. Towards improving the quantity and quality of metadata, we propose a novel metadata prediction framework to learn associations from existing metadata that can be used to predict metadata values. We evaluate our framework in the context of experimental metadata from the Gene Expression Omnibus (GEO). We applied four rule mining algorithms to the most common structured metadata elements (sample type, molecular type, platform, label type and organism) from over 1.3 million GEO records. We examined the quality of well supported rules from each algorithm and visualized the dependencies among metadata elements. Finally, we evaluated the performance of the algorithms in terms of accuracy, precision, recall, and F-measure. We found that PART is the best algorithm outperforming Apriori, Predictive Apriori, and Decision Table.
All algorithms perform significantly better in predicting class values than the majority vote classifier. We found that the performance of the algorithms is related to the dimensionality of the GEO elements. (Full article...)
Recently featured:
- ▪ Rapid development of entity-based data models for bioinformatics with persistence object-oriented design and structured interfaces
- ▪ Bioinformatics education in pathology training: Current scope and future direction
- ▪ FluxCTTX: A LIMS-based tool for management and analysis of cytotoxicity assays data