Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) m (Internal link) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 GroganzOpenSourceBR2011 Aug.png|220px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Benefits of the community for partners of open source vendors|Benefits of the community for partners of open source vendors]]"''' | ||
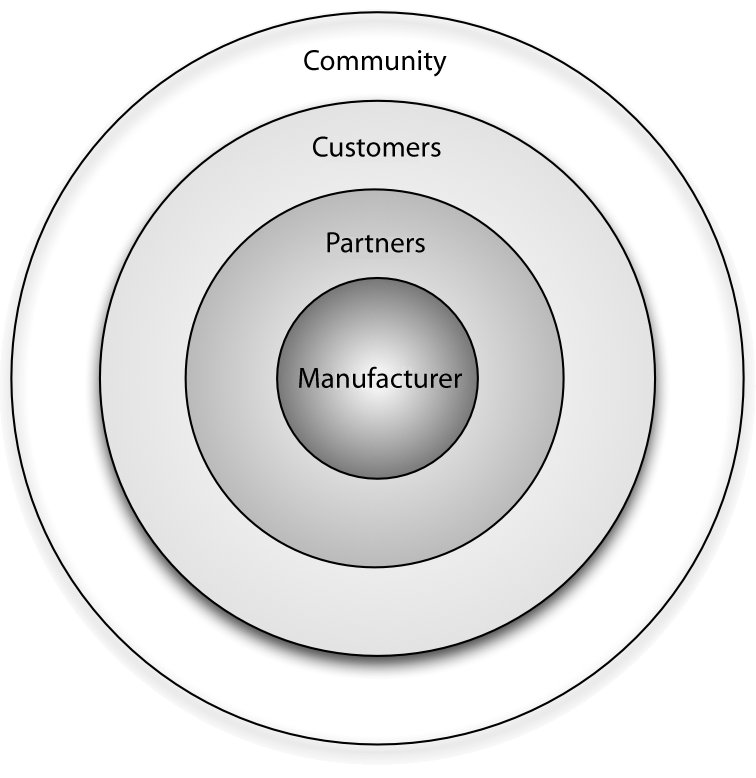
Open source vendors can benefit from business ecosystems that form around their products. Partners of such vendors can utilize this ecosystem for their own business benefit by understanding the structure of the ecosystem, the key actors and their relationships, and the main levers of profitability. This article provides [[information]] on all of these aspects and identifies common business scenarios for partners of open source vendors. Armed with this information, partners can select a strategy that allows them to participate in the ecosystem while also maximizing their gains and driving adoption of their product or solution in the marketplace. | |||
Every [[Free and open-source software#FLOSS|free/libre open source software]] (F/LOSS) vendor strives to create a business ecosystem around its software product. Doing this offers two primary advantages from a sales and marketing perspective: i) it increases the viability and longevity of the product in both commercial and communal spaces, and ii) it opens up new channels for communication and innovation. ('''[[Journal:Benefits of the community for partners of open source vendors|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': [[Journal:MendeLIMS: A web-based laboratory information management system for clinical genome sequencing|MendeLIMS: A web-based laboratory information management system for clinical genome sequencing]], [[Journal:Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages|Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages | ''Recently featured'': [[Journal:adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies|adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies]], [[Journal:MendeLIMS: A web-based laboratory information management system for clinical genome sequencing|MendeLIMS: A web-based laboratory information management system for clinical genome sequencing]], [[Journal:Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages|Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages]] | ||
Revision as of 16:08, 19 January 2016
"Benefits of the community for partners of open source vendors"
Open source vendors can benefit from business ecosystems that form around their products. Partners of such vendors can utilize this ecosystem for their own business benefit by understanding the structure of the ecosystem, the key actors and their relationships, and the main levers of profitability. This article provides information on all of these aspects and identifies common business scenarios for partners of open source vendors. Armed with this information, partners can select a strategy that allows them to participate in the ecosystem while also maximizing their gains and driving adoption of their product or solution in the marketplace.
Every free/libre open source software (F/LOSS) vendor strives to create a business ecosystem around its software product. Doing this offers two primary advantages from a sales and marketing perspective: i) it increases the viability and longevity of the product in both commercial and communal spaces, and ii) it opens up new channels for communication and innovation. (Full article...)
Recently featured: adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies, MendeLIMS: A web-based laboratory information management system for clinical genome sequencing, Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages