Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) m (Italics) |
Shawndouglas (talk | contribs) (Updating article of the week text) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 List ScientificReports2014 4.jpg|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Efficient sample tracking with OpenLabFramework|Efficient sample tracking with OpenLabFramework]]"''' | ||
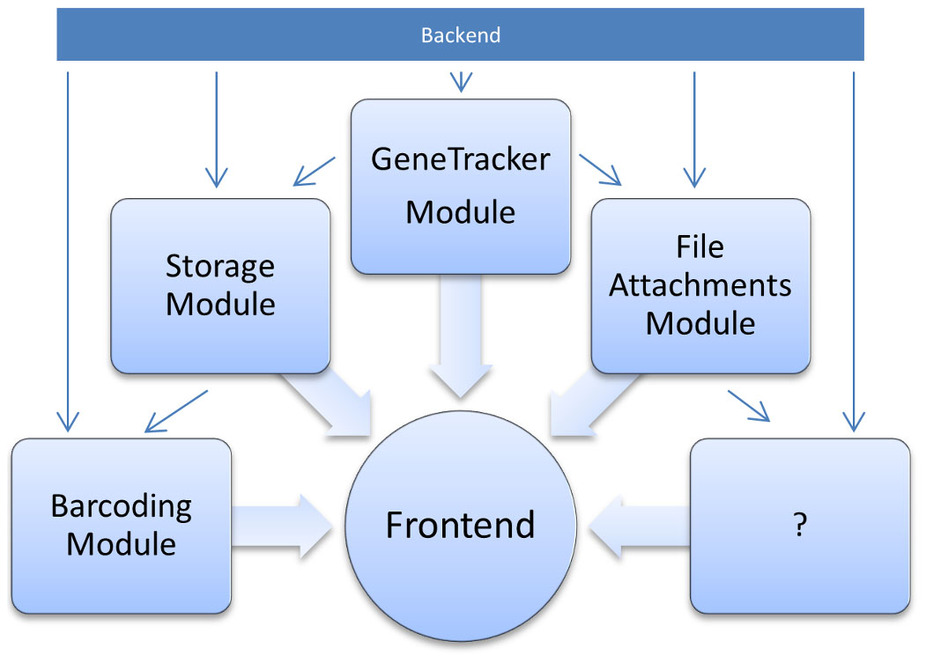
The advance of new technologies in biomedical research has led to a dramatic growth in experimental throughput. Projects therefore steadily grow in size and involve a larger number of researchers. Spreadsheets traditionally used are thus no longer suitable for keeping track of the vast amounts of samples created and need to be replaced with state-of-the-art [[laboratory information management system]]s. Such systems have been developed in large numbers, but they are often limited to specific research domains and types of data. One domain so far neglected is the management of libraries of vector clones and genetically engineered cell lines. [[OpenLabFramework]] is a newly developed web-application for sample tracking, particularly laid out to fill this gap, but with an open architecture allowing it to be extended for other biological materials and functional data. Its sample tracking mechanism is fully customizable and aids productivity further through support for mobile devices and barcoded labels. ('''[[Journal:Efficient sample tracking with OpenLabFramework|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
* [[Journal:Design, implementation and operation of a multimodality research imaging informatics repository|Design, implementation and operation of a multimodality research imaging informatics repository]] | |||
* [[Forensic science]] | * [[Forensic science]] | ||
* [[Journal:Djeen (Database for Joomla!’s Extensible Engine): A research information management system for flexible multi-technology project administration|Djeen (Database for Joomla!’s Extensible Engine): A research information management system for flexible multi-technology project administration]] | * [[Journal:Djeen (Database for Joomla!’s Extensible Engine): A research information management system for flexible multi-technology project administration|Djeen (Database for Joomla!’s Extensible Engine): A research information management system for flexible multi-technology project administration]] | ||
Revision as of 17:15, 21 March 2016
"Efficient sample tracking with OpenLabFramework"
The advance of new technologies in biomedical research has led to a dramatic growth in experimental throughput. Projects therefore steadily grow in size and involve a larger number of researchers. Spreadsheets traditionally used are thus no longer suitable for keeping track of the vast amounts of samples created and need to be replaced with state-of-the-art laboratory information management systems. Such systems have been developed in large numbers, but they are often limited to specific research domains and types of data. One domain so far neglected is the management of libraries of vector clones and genetically engineered cell lines. OpenLabFramework is a newly developed web-application for sample tracking, particularly laid out to fill this gap, but with an open architecture allowing it to be extended for other biological materials and functional data. Its sample tracking mechanism is fully customizable and aids productivity further through support for mobile devices and barcoded labels. (Full article...)
Recently featured: