Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Mariette BMCGenomics2015 13.jpg|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:NG6: Integrated next generation sequencing storage and processing environment|NG6: Integrated next generation sequencing storage and processing environment]]"''' | ||
Next generation [[sequencing]] platforms are now well implanted in sequencing centres and some [[Laboratory|laboratories]]. Upcoming smaller scale machines such as the 454 junior from Roche or the MiSeq from Illumina will increase the number of laboratories hosting a sequencer. In such a context, it is important to provide these teams with an easily manageable environment to store and process the produced reads. | |||
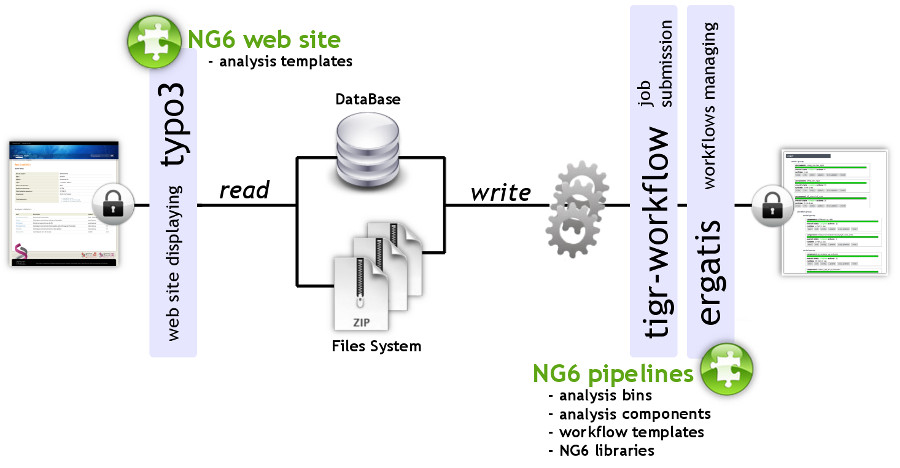
We describe a user-friendly information system able to manage large sets of sequencing data. It includes, on one hand, a workflow environment already containing pipelines adapted to different input formats (sff, fasta, fastq and qseq), different sequencers (Roche 454, Illumina HiSeq) and various analyses (quality control, assembly, alignment, diversity studies,…) and, on the other hand, a secured web site giving access to the results. The connected user will be able to download raw and processed data and browse through the analysis result statistics. ('''[[Journal:NG6: Integrated next generation sequencing storage and processing environment|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:STATegra EMS: An experiment management system for complex next-generation omics experiments|STATegra EMS: An experiment management system for complex next-generation omics experiments]] | |||
: ▪ [[Journal:No specimen left behind: Industrial scale digitization of natural history collections|No specimen left behind: Industrial scale digitization of natural history collections]] | : ▪ [[Journal:No specimen left behind: Industrial scale digitization of natural history collections|No specimen left behind: Industrial scale digitization of natural history collections]] | ||
: ▪ [[Journal:MaPSeq, a service-oriented architecture for genomics research within an academic biomedical research institution|MaPSeq, a service-oriented architecture for genomics research within an academic biomedical research institution]] | : ▪ [[Journal:MaPSeq, a service-oriented architecture for genomics research within an academic biomedical research institution|MaPSeq, a service-oriented architecture for genomics research within an academic biomedical research institution]] | ||
Revision as of 14:40, 9 May 2016
"NG6: Integrated next generation sequencing storage and processing environment"
Next generation sequencing platforms are now well implanted in sequencing centres and some laboratories. Upcoming smaller scale machines such as the 454 junior from Roche or the MiSeq from Illumina will increase the number of laboratories hosting a sequencer. In such a context, it is important to provide these teams with an easily manageable environment to store and process the produced reads.
We describe a user-friendly information system able to manage large sets of sequencing data. It includes, on one hand, a workflow environment already containing pipelines adapted to different input formats (sff, fasta, fastq and qseq), different sequencers (Roche 454, Illumina HiSeq) and various analyses (quality control, assembly, alignment, diversity studies,…) and, on the other hand, a secured web site giving access to the results. The connected user will be able to download raw and processed data and browse through the analysis result statistics. (Full article...)
Recently featured: