Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Vodovnik JofPathInformatics2016 7.jpg|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Diagnostic time in digital pathology: A comparative study on 400 cases|Diagnostic time in digital pathology: A comparative study on 400 cases]]"''' | ||
Numerous validation studies in digital pathology confirmed its value as a diagnostic tool. However, a longer time to diagnosis than traditional microscopy has been seen as a significant barrier to the routine use of digital pathology. As a part of our validation study, we compared a digital and microscopic diagnostic time in the routine diagnostic setting. | |||
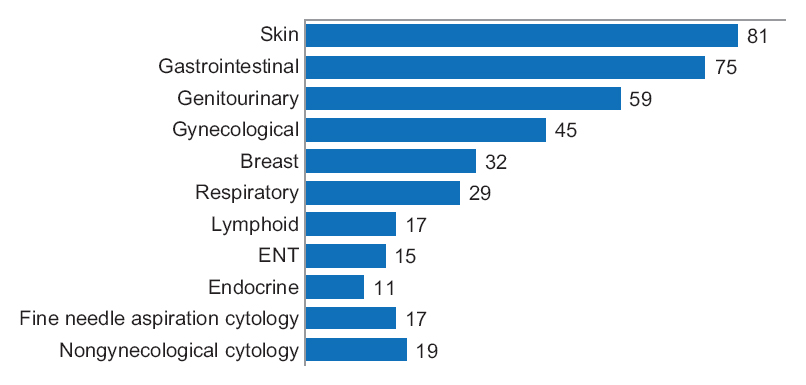
One senior staff pathologist reported 400 consecutive cases in histology, nongynecological, and fine needle aspiration cytology (20 sessions, 20 cases/session), over 4 weeks. Complex, difficult, and rare cases were excluded from the study to reduce the bias. A primary diagnosis was digital, followed by traditional microscopy, six months later, with only request forms available for both. Microscopic slides were scanned at ×20, digital images accessed through the fully integrated [[laboratory information management system]] (LIMS) and viewed in the image viewer on double 23” displays. ('''[[Journal:Diagnostic time in digital pathology: A comparative study on 400 cases|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:OpenChrom: A cross-platform open source software for the mass spectrometric analysis of chromatographic data|OpenChrom: A cross-platform open source software for the mass spectrometric analysis of chromatographic data]] | |||
: ▪ [[Journal:Custom software development for use in a clinical laboratory|Custom software development for use in a clinical laboratory]] | : ▪ [[Journal:Custom software development for use in a clinical laboratory|Custom software development for use in a clinical laboratory]] | ||
: ▪ [[Journal:NG6: Integrated next generation sequencing storage and processing environment|NG6: Integrated next generation sequencing storage and processing environment]] | : ▪ [[Journal:NG6: Integrated next generation sequencing storage and processing environment|NG6: Integrated next generation sequencing storage and processing environment]] | ||
Revision as of 17:28, 31 May 2016
"Diagnostic time in digital pathology: A comparative study on 400 cases"
Numerous validation studies in digital pathology confirmed its value as a diagnostic tool. However, a longer time to diagnosis than traditional microscopy has been seen as a significant barrier to the routine use of digital pathology. As a part of our validation study, we compared a digital and microscopic diagnostic time in the routine diagnostic setting.
One senior staff pathologist reported 400 consecutive cases in histology, nongynecological, and fine needle aspiration cytology (20 sessions, 20 cases/session), over 4 weeks. Complex, difficult, and rare cases were excluded from the study to reduce the bias. A primary diagnosis was digital, followed by traditional microscopy, six months later, with only request forms available for both. Microscopic slides were scanned at ×20, digital images accessed through the fully integrated laboratory information management system (LIMS) and viewed in the image viewer on double 23” displays. (Full article...)
Recently featured: