Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Maier MammalianGenome2015 26-9.gif|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Principles and application of LIMS in mouse clinics|Principles and application of LIMS in mouse clinics]]"''' | ||
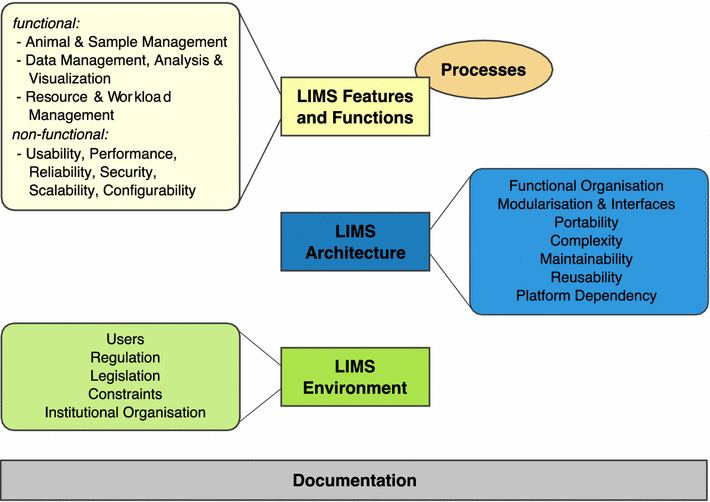
Large-scale systemic mouse phenotyping, as performed by mouse clinics for more than a decade, requires thousands of mice from a multitude of different mutant lines to be bred, individually tracked and subjected to phenotyping procedures according to a standardised schedule. All these efforts are typically organised in overlapping projects, running in parallel. In terms of logistics, data capture, [[data analysis]], result visualisation and reporting, new challenges have emerged from such projects. These challenges could hardly be met with traditional methods such as pen and paper colony management, spreadsheet-based data management and manual data analysis. Hence, different [[laboratory information management system]]s (LIMS) have been developed in mouse clinics to facilitate or even enable mouse and data management in the described order of magnitude. This review shows that general principles of LIMS can be empirically deduced from LIMS used by different mouse clinics, although these have evolved differently. Supported by LIMS descriptions and lessons learned from seven mouse clinics, this review also shows that the unique LIMS environment in a particular facility strongly influences strategic LIMS decisions and LIMS development. As a major conclusion, this review states that there is no universal LIMS for the mouse research domain that fits all requirements. Still, empirically deduced general LIMS principles can serve as a master decision support template, which is provided as a hands-on tool for mouse research facilities looking for a LIMS. ('''[[Journal:Principles and application of LIMS in mouse clinics|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Multilevel classification of security concerns in cloud computing|Multilevel classification of security concerns in cloud computing]] | |||
: ▪ [[Journal:Assessment of and response to data needs of clinical and translational science researchers and beyond|Assessment of and response to data needs of clinical and translational science researchers and beyond]] | : ▪ [[Journal:Assessment of and response to data needs of clinical and translational science researchers and beyond|Assessment of and response to data needs of clinical and translational science researchers and beyond]] | ||
: ▪ [[Journal:SUSHI: An exquisite recipe for fully documented, reproducible and reusable NGS data analysis|SUSHI: An exquisite recipe for fully documented, reproducible and reusable NGS data analysis]] | : ▪ [[Journal:SUSHI: An exquisite recipe for fully documented, reproducible and reusable NGS data analysis|SUSHI: An exquisite recipe for fully documented, reproducible and reusable NGS data analysis]] | ||
Revision as of 16:00, 26 September 2016
"Principles and application of LIMS in mouse clinics"
Large-scale systemic mouse phenotyping, as performed by mouse clinics for more than a decade, requires thousands of mice from a multitude of different mutant lines to be bred, individually tracked and subjected to phenotyping procedures according to a standardised schedule. All these efforts are typically organised in overlapping projects, running in parallel. In terms of logistics, data capture, data analysis, result visualisation and reporting, new challenges have emerged from such projects. These challenges could hardly be met with traditional methods such as pen and paper colony management, spreadsheet-based data management and manual data analysis. Hence, different laboratory information management systems (LIMS) have been developed in mouse clinics to facilitate or even enable mouse and data management in the described order of magnitude. This review shows that general principles of LIMS can be empirically deduced from LIMS used by different mouse clinics, although these have evolved differently. Supported by LIMS descriptions and lessons learned from seven mouse clinics, this review also shows that the unique LIMS environment in a particular facility strongly influences strategic LIMS decisions and LIMS development. As a major conclusion, this review states that there is no universal LIMS for the mouse research domain that fits all requirements. Still, empirically deduced general LIMS principles can serve as a master decision support template, which is provided as a hands-on tool for mouse research facilities looking for a LIMS. (Full article...)
Recently featured: