Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Naulaerts BioAndBioInsights2016 10.png|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Practical approaches for mining frequent patterns in molecular datasets|Practical approaches for mining frequent patterns in molecular datasets]]"''' | ||
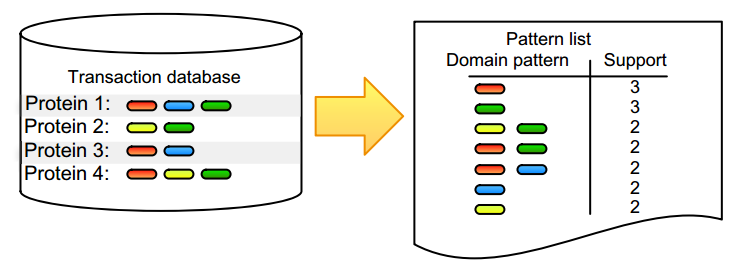
Pattern detection is an inherent task in the analysis and interpretation of complex and continuously accumulating biological data. Numerous [[wikipedia:Sequential pattern mining|itemset mining]] algorithms have been developed in the last decade to efficiently detect specific pattern classes in data. Although many of these have proven their value for addressing bioinformatics problems, several factors still slow down promising algorithms from gaining popularity in the life science community. Many of these issues stem from the low user-friendliness of these tools and the complexity of their output, which is often large, static, and consequently hard to interpret. Here, we apply three software implementations on common [[bioinformatics]] problems and illustrate some of the advantages and disadvantages of each, as well as inherent pitfalls of biological data mining. Frequent itemset mining exists in many different flavors, and users should decide their software choice based on their research question, programming proficiency, and added value of extra features. ('''[[Journal:Practical approaches for mining frequent patterns in molecular datasets|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Improving the creation and reporting of structured findings during digital pathology review|Improving the creation and reporting of structured findings during digital pathology review]] | |||
: ▪ [[Journal:The challenges of data quality and data quality assessment in the big data era|The challenges of data quality and data quality assessment in the big data era]] | : ▪ [[Journal:The challenges of data quality and data quality assessment in the big data era|The challenges of data quality and data quality assessment in the big data era]] | ||
: ▪ [[Journal:Water, water, everywhere: Defining and assessing data sharing in academia|Water, water, everywhere: Defining and assessing data sharing in academia]] | : ▪ [[Journal:Water, water, everywhere: Defining and assessing data sharing in academia|Water, water, everywhere: Defining and assessing data sharing in academia]] | ||
Revision as of 17:59, 25 October 2016
"Practical approaches for mining frequent patterns in molecular datasets"
Pattern detection is an inherent task in the analysis and interpretation of complex and continuously accumulating biological data. Numerous itemset mining algorithms have been developed in the last decade to efficiently detect specific pattern classes in data. Although many of these have proven their value for addressing bioinformatics problems, several factors still slow down promising algorithms from gaining popularity in the life science community. Many of these issues stem from the low user-friendliness of these tools and the complexity of their output, which is often large, static, and consequently hard to interpret. Here, we apply three software implementations on common bioinformatics problems and illustrate some of the advantages and disadvantages of each, as well as inherent pitfalls of biological data mining. Frequent itemset mining exists in many different flavors, and users should decide their software choice based on their research question, programming proficiency, and added value of extra features. (Full article...)
Recently featured: