Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Goldberg mBio2015 6-6.jpg|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Making the leap from research laboratory to clinic: Challenges and opportunities for next-generation sequencing in infectious disease diagnostics|Making the leap from research laboratory to clinic: Challenges and opportunities for next-generation sequencing in infectious disease diagnostics]]"''' | ||
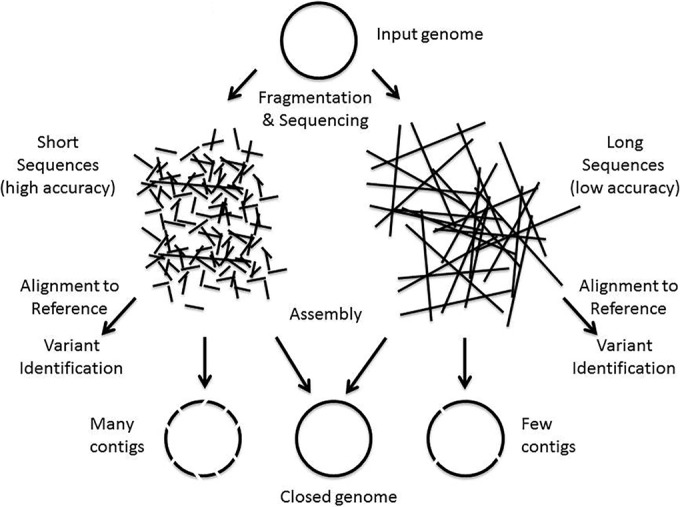
Next-generation DNA sequencing (NGS) has progressed enormously over the past decade, transforming [[Genomics|genomic]] analysis and opening up many new opportunities for applications in clinical microbiology [[Laboratory|laboratories]]. The impact of NGS on microbiology has been revolutionary, with new microbial genomic sequences being generated daily, leading to the development of large databases of genomes and gene sequences. The ability to analyze microbial communities without culturing organisms has created the ever-growing field of metagenomics and microbiome analysis and has generated significant new insights into the relation between host and microbe. The medical literature contains many examples of how this new technology can be used for infectious disease diagnostics and pathogen analysis. The implementation of NGS in medical practice has been a slow process due to various challenges such as clinical trials, lack of applicable regulatory guidelines, and the adaptation of the technology to the clinical environment. In April 2015, the American Academy of Microbiology (AAM) convened a colloquium to begin to define these issues, and in this document, we present some of the concepts that were generated from these discussions. ('''[[Journal:Making the leap from research laboratory to clinic: Challenges and opportunities for next-generation sequencing in infectious disease diagnostics|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Information technology support for clinical genetic testing within an academic medical center|Information technology support for clinical genetic testing within an academic medical center]] | |||
: ▪ [[Journal:Principles of metadata organization at the ENCODE data coordination center|Principles of metadata organization at the ENCODE data coordination center]] | : ▪ [[Journal:Principles of metadata organization at the ENCODE data coordination center|Principles of metadata organization at the ENCODE data coordination center]] | ||
: ▪ [[Journal:Integrated systems for NGS data management and analysis: Open issues and available solutions|Integrated systems for NGS data management and analysis: Open issues and available solutions]] | : ▪ [[Journal:Integrated systems for NGS data management and analysis: Open issues and available solutions|Integrated systems for NGS data management and analysis: Open issues and available solutions]] | ||
Revision as of 15:51, 21 November 2016
Next-generation DNA sequencing (NGS) has progressed enormously over the past decade, transforming genomic analysis and opening up many new opportunities for applications in clinical microbiology laboratories. The impact of NGS on microbiology has been revolutionary, with new microbial genomic sequences being generated daily, leading to the development of large databases of genomes and gene sequences. The ability to analyze microbial communities without culturing organisms has created the ever-growing field of metagenomics and microbiome analysis and has generated significant new insights into the relation between host and microbe. The medical literature contains many examples of how this new technology can be used for infectious disease diagnostics and pathogen analysis. The implementation of NGS in medical practice has been a slow process due to various challenges such as clinical trials, lack of applicable regulatory guidelines, and the adaptation of the technology to the clinical environment. In April 2015, the American Academy of Microbiology (AAM) convened a colloquium to begin to define these issues, and in this document, we present some of the concepts that were generated from these discussions. (Full article...)
Recently featured: