Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Woinarowicz OJPHI2016 8-2.png|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:The impact of electronic health record (EHR) interoperability on immunization information system (IIS) data quality|The impact of electronic health record (EHR) interoperability on immunization information system (IIS) data quality]]"''' | ||
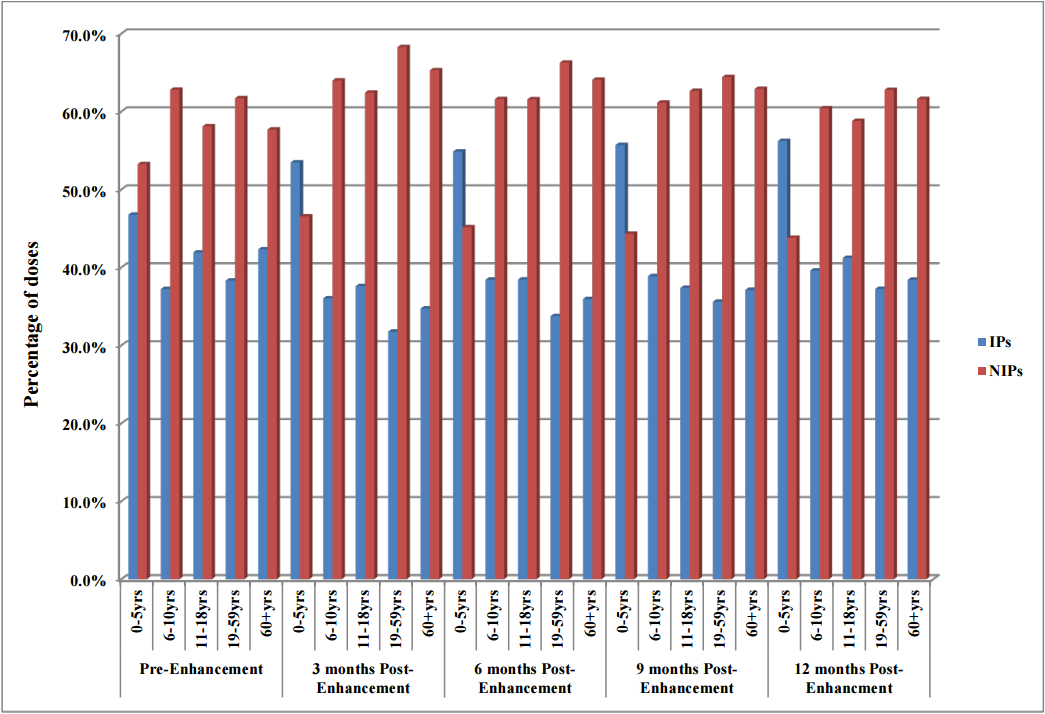
Objectives: To evaluate the impact of [[electronic health record]] (EHR) interoperability on the quality of immunization data in the North Dakota Immunization Information System (NDIIS). | |||
NDIIS "doses administered" data was evaluated for completeness of the patient and dose-level core data elements for records that belong to interoperable and non-interoperable providers. Data was compared at three months prior to EHR interoperability enhancement to data at three, six, nine and 12 months post-enhancement following the interoperability go live date. Doses administered per month and by age group, timeliness of vaccine entry and the number of duplicate clients added to the NDIIS was also compared, in addition to immunization rates for children 19–35 months of age and adolescents 11–18 years of age. ('''[[Journal:The impact of electronic health record (EHR) interoperability on immunization information system (IIS) data quality|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Bioinformatics workflow for clinical whole genome sequencing at Partners HealthCare Personalized Medicine|Bioinformatics workflow for clinical whole genome sequencing at Partners HealthCare Personalized Medicine]] | |||
: ▪ [[Journal:Pathology report data extraction from relational database using R, with extraction from reports on melanoma of skin as an example|Pathology report data extraction from relational database using R, with extraction from reports on melanoma of skin as an example]] | : ▪ [[Journal:Pathology report data extraction from relational database using R, with extraction from reports on melanoma of skin as an example|Pathology report data extraction from relational database using R, with extraction from reports on melanoma of skin as an example]] | ||
: ▪ [[Journal:A robust, format-agnostic scientific data transfer framework|A robust, format-agnostic scientific data transfer framework]] | : ▪ [[Journal:A robust, format-agnostic scientific data transfer framework|A robust, format-agnostic scientific data transfer framework]] | ||
Revision as of 16:09, 7 February 2017
Objectives: To evaluate the impact of electronic health record (EHR) interoperability on the quality of immunization data in the North Dakota Immunization Information System (NDIIS).
NDIIS "doses administered" data was evaluated for completeness of the patient and dose-level core data elements for records that belong to interoperable and non-interoperable providers. Data was compared at three months prior to EHR interoperability enhancement to data at three, six, nine and 12 months post-enhancement following the interoperability go live date. Doses administered per month and by age group, timeliness of vaccine entry and the number of duplicate clients added to the NDIIS was also compared, in addition to immunization rates for children 19–35 months of age and adolescents 11–18 years of age. (Full article...)
Recently featured:
- ▪ Bioinformatics workflow for clinical whole genome sequencing at Partners HealthCare Personalized Medicine
- ▪ Pathology report data extraction from relational database using R, with extraction from reports on melanoma of skin as an example
- ▪ A robust, format-agnostic scientific data transfer framework