Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Schulz JofPathInformatics2016 7.jpg|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Use of application containers and workflows for genomic data analysis|Use of application containers and workflows for genomic data analysis]]"''' | ||
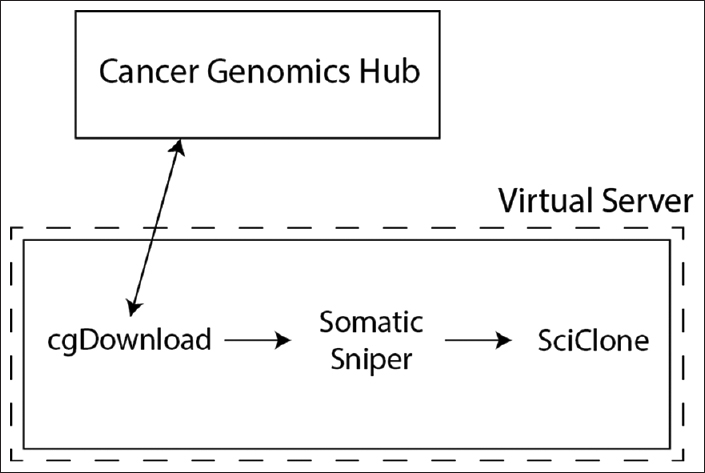
[[ | The rapid acquisition of biological data and development of computationally intensive analyses has led to a need for novel approaches to software deployment. In particular, the complexity of common analytic tools for [[genomics]] makes them difficult to deploy and decreases the reproducibility of computational experiments. Recent technologies that allow for application virtualization, such as Docker, allow developers and bioinformaticians to isolate these applications and deploy secure, scalable platforms that have the potential to dramatically increase the efficiency of big data processing. While limitations exist, this study demonstrates a successful implementation of a pipeline with several discrete software applications for the analysis of next-generation sequencing (NGS) data. ('''[[Journal:Use of application containers and workflows for genomic data analysis|Full article...]]''')<br /> | ||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Informatics metrics and measures for a smart public health systems approach: Information science perspective|Informatics metrics and measures for a smart public health systems approach: Information science perspective]] | |||
: ▪ [[Journal:Deployment of analytics into the healthcare safety net: Lessons learned|Deployment of analytics into the healthcare safety net: Lessons learned]] | : ▪ [[Journal:Deployment of analytics into the healthcare safety net: Lessons learned|Deployment of analytics into the healthcare safety net: Lessons learned]] | ||
: ▪ [[Journal:The growing need for microservices in bioinformatics|The growing need for microservices in bioinformatics]] | : ▪ [[Journal:The growing need for microservices in bioinformatics|The growing need for microservices in bioinformatics]] | ||
Revision as of 16:41, 13 March 2017
"Use of application containers and workflows for genomic data analysis"
The rapid acquisition of biological data and development of computationally intensive analyses has led to a need for novel approaches to software deployment. In particular, the complexity of common analytic tools for genomics makes them difficult to deploy and decreases the reproducibility of computational experiments. Recent technologies that allow for application virtualization, such as Docker, allow developers and bioinformaticians to isolate these applications and deploy secure, scalable platforms that have the potential to dramatically increase the efficiency of big data processing. While limitations exist, this study demonstrates a successful implementation of a pipeline with several discrete software applications for the analysis of next-generation sequencing (NGS) data. (Full article...)
Recently featured: