Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) m (Image) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig5 Lukauskas BMCBioinformatics2016 17-Supp16.gif|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:DGW: An exploratory data analysis tool for clustering and visualisation of epigenomic marks|DGW: An exploratory data analysis tool for clustering and visualisation of epigenomic marks]]"''' | ||
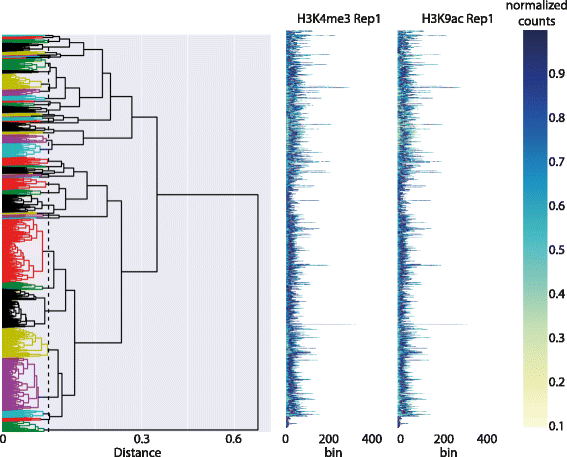
Functional [[Genomics|genomic]] and epigenomic research relies fundamentally on [[sequencing]]-based methods like ChIP-seq for the detection of DNA-protein interactions. These techniques return large, high-dimensional data sets with visually complex structures, such as multi-modal peaks extended over large genomic regions. Current tools for visualisation and data exploration represent and leverage these complex features only to a limited extent. | |||
We present DGW (Dynamic Gene Warping), an open-source software package for simultaneous alignment and clustering of multiple epigenomic marks. DGW uses dynamic time warping to adaptively rescale and align genomic distances which allows to group regions of interest with similar shapes, thereby capturing the structure of epigenomic marks. We demonstrate the effectiveness of the approach in a simulation study and on a real epigenomic data set from the ENCODE project. ('''[[Journal:DGW: An exploratory data analysis tool for clustering and visualisation of epigenomic marks|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:SCIFIO: An extensible framework to support scientific image formats|SCIFIO: An extensible framework to support scientific image formats]] | |||
: ▪ [[Journal:Use of application containers and workflows for genomic data analysis|Use of application containers and workflows for genomic data analysis]] | : ▪ [[Journal:Use of application containers and workflows for genomic data analysis|Use of application containers and workflows for genomic data analysis]] | ||
: ▪ [[Journal:Informatics metrics and measures for a smart public health systems approach: Information science perspective|Informatics metrics and measures for a smart public health systems approach: Information science perspective]] | : ▪ [[Journal:Informatics metrics and measures for a smart public health systems approach: Information science perspective|Informatics metrics and measures for a smart public health systems approach: Information science perspective]] | ||
Revision as of 15:37, 27 March 2017
"DGW: An exploratory data analysis tool for clustering and visualisation of epigenomic marks"
Functional genomic and epigenomic research relies fundamentally on sequencing-based methods like ChIP-seq for the detection of DNA-protein interactions. These techniques return large, high-dimensional data sets with visually complex structures, such as multi-modal peaks extended over large genomic regions. Current tools for visualisation and data exploration represent and leverage these complex features only to a limited extent.
We present DGW (Dynamic Gene Warping), an open-source software package for simultaneous alignment and clustering of multiple epigenomic marks. DGW uses dynamic time warping to adaptively rescale and align genomic distances which allows to group regions of interest with similar shapes, thereby capturing the structure of epigenomic marks. We demonstrate the effectiveness of the approach in a simulation study and on a real epigenomic data set from the ENCODE project. (Full article...)
Recently featured: