Difference between revisions of "Template:Article of the week"
From LIMSWiki
Jump to navigationJump to searchShawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Harvey JoCheminformatics2017 9.gif|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:A metadata-driven approach to data repository design|A metadata-driven approach to data repository design]]"''' | ||
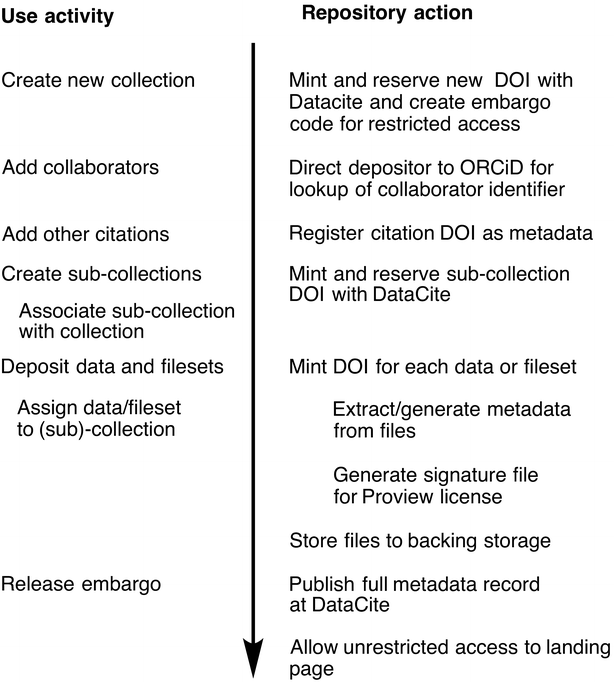
The design and use of a metadata-driven data repository for research data management is described. Metadata is collected automatically during the submission process whenever possible and is registered with DataCite in accordance with their current metadata schema, in exchange for a persistent digital object identifier. Two examples of data preview are illustrated, including the demonstration of a method for integration with commercial software that confers rich domain-specific [[data analysis|data analytics]] without introducing customization into the repository itself. ('''[[Journal:A metadata-driven approach to data repository design|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:DGW: An exploratory data analysis tool for clustering and visualisation of epigenomic marks|DGW: An exploratory data analysis tool for clustering and visualisation of epigenomic marks]] | |||
: ▪ [[Journal:SCIFIO: An extensible framework to support scientific image formats|SCIFIO: An extensible framework to support scientific image formats]] | : ▪ [[Journal:SCIFIO: An extensible framework to support scientific image formats|SCIFIO: An extensible framework to support scientific image formats]] | ||
: ▪ [[Journal:Use of application containers and workflows for genomic data analysis|Use of application containers and workflows for genomic data analysis]] | : ▪ [[Journal:Use of application containers and workflows for genomic data analysis|Use of application containers and workflows for genomic data analysis]] | ||
Revision as of 19:15, 3 April 2017
"A metadata-driven approach to data repository design"
The design and use of a metadata-driven data repository for research data management is described. Metadata is collected automatically during the submission process whenever possible and is registered with DataCite in accordance with their current metadata schema, in exchange for a persistent digital object identifier. Two examples of data preview are illustrated, including the demonstration of a method for integration with commercial software that confers rich domain-specific data analytics without introducing customization into the repository itself. (Full article...)
Recently featured: