Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Rubel FInNeuroinformatics2016 10.jpg|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Methods for specifying scientific data standards and modeling relationships with applications to neuroscience|Methods for specifying scientific data standards and modeling relationships with applications to neuroscience]]"''' | ||
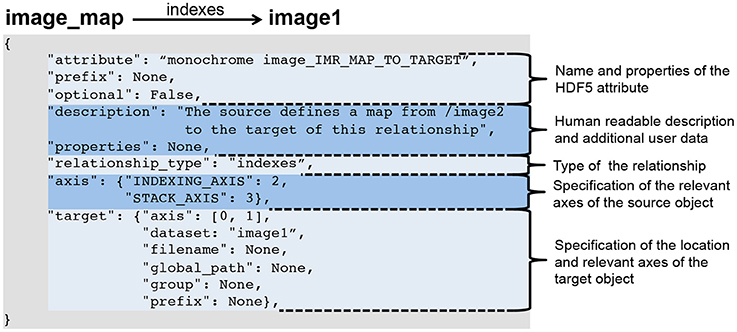
Neuroscience continues to experience a tremendous growth in data; in terms of the volume and variety of data, the velocity at which data is acquired, and in turn the veracity of data. These challenges are a serious impediment to sharing of data, analyses, and tools within and across labs. Here, we introduce BRAINformat, a novel data standardization framework for the design and management of scientific data formats. The BRAINformat library defines application-independent design concepts and modules that together create a general framework for standardization of scientific data. We describe the formal specification of scientific data standards, which facilitates sharing and verification of data and formats. We introduce the concept of "managed objects," enabling semantic components of data formats to be specified as self-contained units, supporting modular and reusable design of data format components and file storage. We also introduce the novel concept of "relationship attributes" for modeling and use of semantic relationships between data objects. Based on these concepts we demonstrate the application of our framework to design and implement a standard format for electrophysiology data and show how data standardization and relationship-modeling facilitate [[data analysis]] and sharing. ('''[[Journal:Methods for specifying scientific data standards and modeling relationships with applications to neuroscience|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Data and metadata brokering – Theory and practice from the BCube Project|Data and metadata brokering – Theory and practice from the BCube Project]] | |||
: ▪ [[Journal:A metadata-driven approach to data repository design|A metadata-driven approach to data repository design]] | : ▪ [[Journal:A metadata-driven approach to data repository design|A metadata-driven approach to data repository design]] | ||
: ▪ [[Journal:DGW: An exploratory data analysis tool for clustering and visualisation of epigenomic marks|DGW: An exploratory data analysis tool for clustering and visualisation of epigenomic marks]] | : ▪ [[Journal:DGW: An exploratory data analysis tool for clustering and visualisation of epigenomic marks|DGW: An exploratory data analysis tool for clustering and visualisation of epigenomic marks]] | ||
Revision as of 15:42, 17 April 2017
Neuroscience continues to experience a tremendous growth in data; in terms of the volume and variety of data, the velocity at which data is acquired, and in turn the veracity of data. These challenges are a serious impediment to sharing of data, analyses, and tools within and across labs. Here, we introduce BRAINformat, a novel data standardization framework for the design and management of scientific data formats. The BRAINformat library defines application-independent design concepts and modules that together create a general framework for standardization of scientific data. We describe the formal specification of scientific data standards, which facilitates sharing and verification of data and formats. We introduce the concept of "managed objects," enabling semantic components of data formats to be specified as self-contained units, supporting modular and reusable design of data format components and file storage. We also introduce the novel concept of "relationship attributes" for modeling and use of semantic relationships between data objects. Based on these concepts we demonstrate the application of our framework to design and implement a standard format for electrophysiology data and show how data standardization and relationship-modeling facilitate data analysis and sharing. (Full article...)
Recently featured: