Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
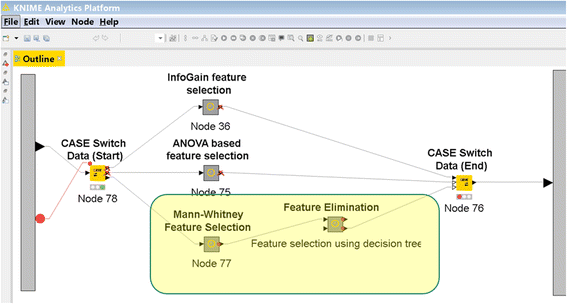
'''"[[Journal: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Eyal-Altman BMCBioinformatics2017 18.gif|240px]]</div> | ||
'''"[[Journal:PCM-SABRE: A platform for benchmarking and comparing outcome prediction methods in precision cancer medicine|PCM-SABRE: A platform for benchmarking and comparing outcome prediction methods in precision cancer medicine]]"''' | |||
Numerous publications attempt to predict cancer survival outcome from gene expression data using machine-learning methods. A direct comparison of these works is challenging for the following reasons: (1) inconsistent measures used to evaluate the performance of different models, and (2) incomplete specification of critical stages in the process of knowledge discovery. There is a need for a platform that would allow researchers to replicate previous works and to test the impact of changes in the knowledge discovery process on the accuracy of the induced models. | |||
We developed the PCM-SABRE platform, which supports the entire knowledge discovery process for cancer outcome analysis. PCM-SABRE was developed using [[KNIME]]. By using PCM-SABRE to reproduce the results of previously published works on breast cancer survival, we define a baseline for evaluating future attempts to predict cancer outcome with machine learning. ('''[[PCM-SABRE: A platform for benchmarking and comparing outcome prediction methods in precision cancer medicine|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Ten simple rules for cultivating open science and collaborative R&D|Ten simple rules for cultivating open science and collaborative R&D]] | |||
: ▪ [[Journal:Ten simple rules to enable multi-site collaborations through data sharing|Ten simple rules to enable multi-site collaborations through data sharing]] | : ▪ [[Journal:Ten simple rules to enable multi-site collaborations through data sharing|Ten simple rules to enable multi-site collaborations through data sharing]] | ||
: ▪ [[Journal:Ten simple rules for developing usable software in computational biology|Ten simple rules for developing usable software in computational biology]] | : ▪ [[Journal:Ten simple rules for developing usable software in computational biology|Ten simple rules for developing usable software in computational biology]] | ||
Revision as of 16:11, 22 May 2017
Numerous publications attempt to predict cancer survival outcome from gene expression data using machine-learning methods. A direct comparison of these works is challenging for the following reasons: (1) inconsistent measures used to evaluate the performance of different models, and (2) incomplete specification of critical stages in the process of knowledge discovery. There is a need for a platform that would allow researchers to replicate previous works and to test the impact of changes in the knowledge discovery process on the accuracy of the induced models.
We developed the PCM-SABRE platform, which supports the entire knowledge discovery process for cancer outcome analysis. PCM-SABRE was developed using KNIME. By using PCM-SABRE to reproduce the results of previously published works on breast cancer survival, we define a baseline for evaluating future attempts to predict cancer outcome with machine learning. (Full article...)
Recently featured: