Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Tab2 Albert AfricanJofLabMed2017 6-2.jpg|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Developing a customized approach for strengthening tuberculosis laboratory quality management systems toward accreditation|Developing a customized approach for strengthening tuberculosis laboratory quality management systems toward accreditation]]"''' | ||
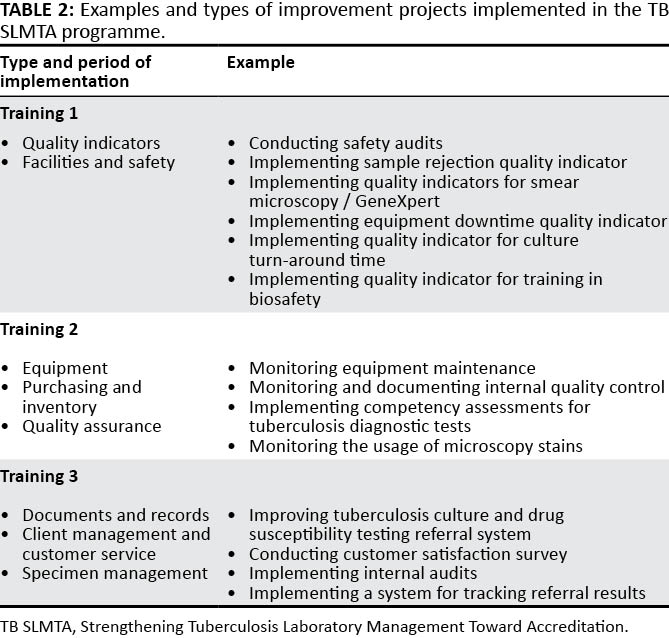
Quality-assured tuberculosis [[laboratory]] services are critical to achieve global and national goals for tuberculosis prevention and care. Implementation of a [[quality management system]] (QMS) in laboratories leads to improved quality of diagnostic tests and better patient care. The Strengthening Laboratory Management Toward Accreditation (SLMTA) program has led to measurable improvements in the QMS of [[clinical laboratory|clinical laboratories]]. However, progress in tuberculosis laboratories has been slower, which may be attributed to the need for a structured tuberculosis-specific approach to implementing QMS. We describe the development and early implementation of the Strengthening Tuberculosis Laboratory Management Toward Accreditation (TB SLMTA) program. The TB SLMTA curriculum was developed by customizing the SLMTA curriculum to include specific tools, job aids, and supplementary materials specific to the tuberculosis laboratory. ('''[[Journal:Developing a customized approach for strengthening tuberculosis laboratory quality management systems toward accreditation|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:SistematX, an online web-based cheminformatics tool for data management of secondary metabolites|SistematX, an online web-based cheminformatics tool for data management of secondary metabolites]] | |||
: ▪ [[Journal:Rethinking data sharing and human participant protection in social science research: Applications from the qualitative realm|Rethinking data sharing and human participant protection in social science research: Applications from the qualitative realm]] | : ▪ [[Journal:Rethinking data sharing and human participant protection in social science research: Applications from the qualitative realm|Rethinking data sharing and human participant protection in social science research: Applications from the qualitative realm]] | ||
: ▪ [[Journal:Handling metadata in a neurophysiology laboratory|Handling metadata in a neurophysiology laboratory]] | : ▪ [[Journal:Handling metadata in a neurophysiology laboratory|Handling metadata in a neurophysiology laboratory]] | ||
Revision as of 21:06, 5 March 2018
Quality-assured tuberculosis laboratory services are critical to achieve global and national goals for tuberculosis prevention and care. Implementation of a quality management system (QMS) in laboratories leads to improved quality of diagnostic tests and better patient care. The Strengthening Laboratory Management Toward Accreditation (SLMTA) program has led to measurable improvements in the QMS of clinical laboratories. However, progress in tuberculosis laboratories has been slower, which may be attributed to the need for a structured tuberculosis-specific approach to implementing QMS. We describe the development and early implementation of the Strengthening Tuberculosis Laboratory Management Toward Accreditation (TB SLMTA) program. The TB SLMTA curriculum was developed by customizing the SLMTA curriculum to include specific tools, job aids, and supplementary materials specific to the tuberculosis laboratory. (Full article...)
Recently featured: