Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
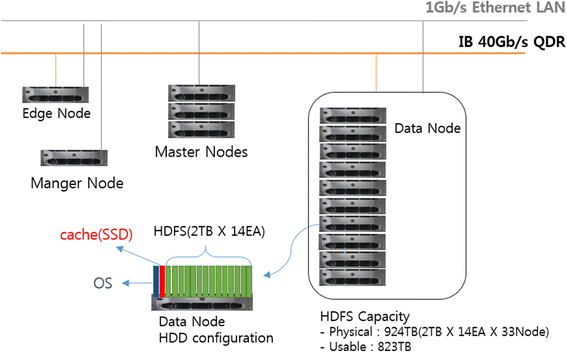
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Closha BMCBioinfo2018 19-Sup1.gif|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Closha: Bioinformatics workflow system for the analysis of massive sequencing data|Closha: Bioinformatics workflow system for the analysis of massive sequencing data]]"''' | ||
While next-generation sequencing (NGS) costs have fallen in recent years, the cost and complexity of computation remain substantial obstacles to the use of NGS in bio-medical care and [[Genomics|genomic]] research. The rapidly increasing amounts of data available from the new high-throughput methods have made data processing infeasible without automated pipelines. The integration of data and analytic resources into workflow systems provides a solution to the problem by simplifying the task of data analysis. | |||
To address this challenge, we developed a cloud-based workflow management system, Closha, to provide fast and cost-effective analysis of massive genomic data. We implemented complex workflows making optimal use of high-performance computing clusters. Closha allows users to create multi-step analyses using drag-and-drop functionality and to modify the parameters of pipeline tools. ('''[[Journal:Closha: Bioinformatics workflow system for the analysis of massive sequencing data|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Big data management for cloud-enabled geological information services|Big data management for cloud-enabled geological information services]] | |||
: ▪ [[Journal:Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory|Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory]] | : ▪ [[Journal:Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory|Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory]] | ||
: ▪ [[Journal:Moving ERP systems to the cloud: Data security issues|Moving ERP systems to the cloud: Data security issues]] | : ▪ [[Journal:Moving ERP systems to the cloud: Data security issues|Moving ERP systems to the cloud: Data security issues]] | ||
Revision as of 15:34, 16 April 2018
"Closha: Bioinformatics workflow system for the analysis of massive sequencing data"
While next-generation sequencing (NGS) costs have fallen in recent years, the cost and complexity of computation remain substantial obstacles to the use of NGS in bio-medical care and genomic research. The rapidly increasing amounts of data available from the new high-throughput methods have made data processing infeasible without automated pipelines. The integration of data and analytic resources into workflow systems provides a solution to the problem by simplifying the task of data analysis.
To address this challenge, we developed a cloud-based workflow management system, Closha, to provide fast and cost-effective analysis of massive genomic data. We implemented complex workflows making optimal use of high-performance computing clusters. Closha allows users to create multi-step analyses using drag-and-drop functionality and to modify the parameters of pipeline tools. (Full article...)
Recently featured: