Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Beaulieu-JonesJMIRMedInfo2018 6-1.png|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Characterizing and managing missing structured data in electronic health records: Data analysis|Characterizing and managing missing structured data in electronic health records: Data analysis]]"''' | ||
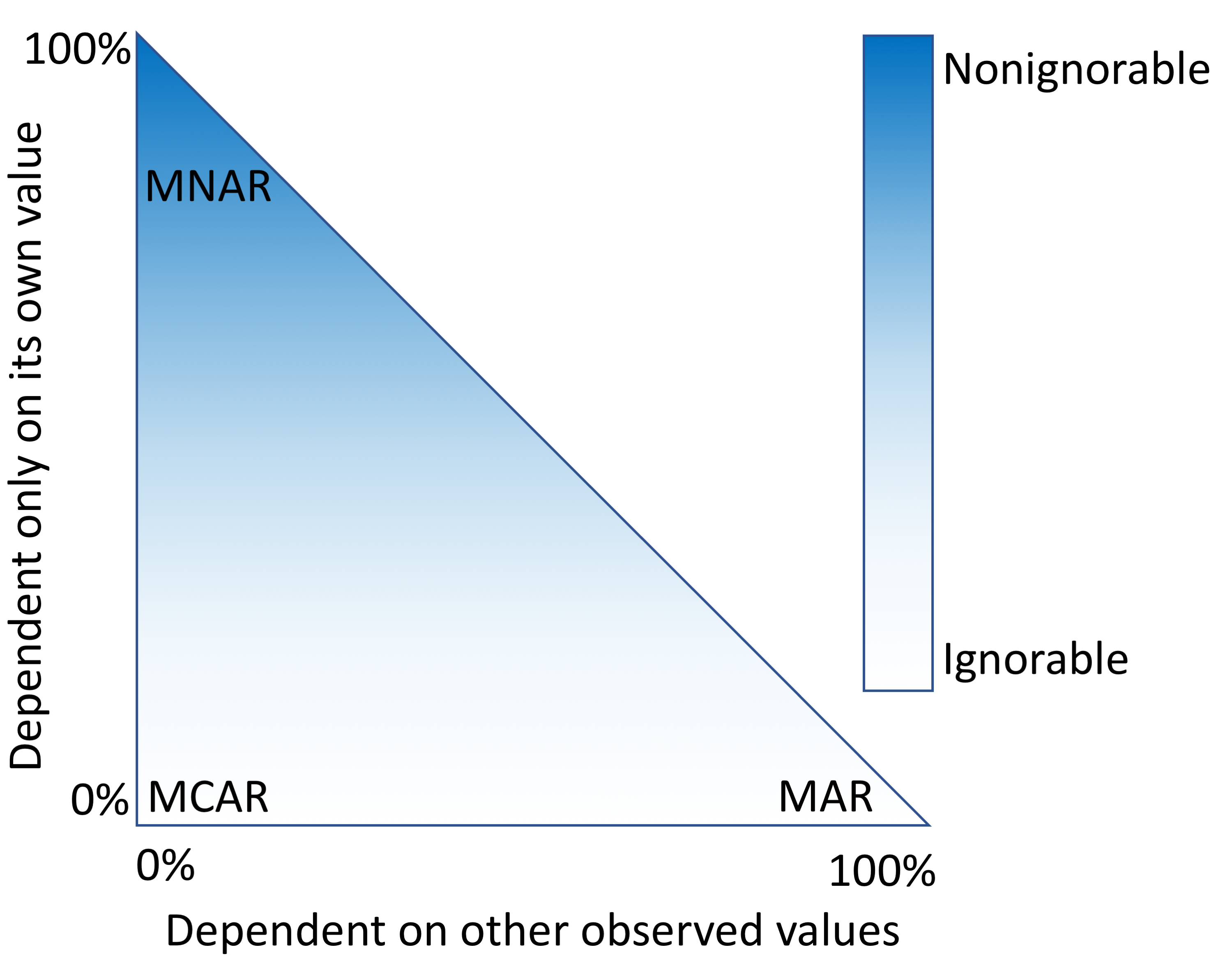
Missing data is a challenge for all studies; however, this is especially true for [[electronic health record]] (EHR)-based analyses. Failure to appropriately consider missing data can lead to biased results. While there has been extensive theoretical work on imputation, and many sophisticated methods are now available, it remains quite challenging for researchers to implement these methods appropriately. Here, we provide detailed procedures for when and how to conduct imputation of EHR [[laboratory]] results. | |||
The objective of this study was to demonstrate how the mechanism of "missingness" can be assessed, evaluate the performance of a variety of imputation methods, and describe some of the most frequent problems that can be encountered. ('''[[Journal:Characterizing and managing missing structured data in electronic health records: Data analysis|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Closha: Bioinformatics workflow system for the analysis of massive sequencing data|Closha: Bioinformatics workflow system for the analysis of massive sequencing data]] | |||
: ▪ [[Journal:Big data management for cloud-enabled geological information services|Big data management for cloud-enabled geological information services]] | : ▪ [[Journal:Big data management for cloud-enabled geological information services|Big data management for cloud-enabled geological information services]] | ||
: ▪ [[Journal:Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory|Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory]] | : ▪ [[Journal:Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory|Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory]] | ||
Revision as of 15:36, 23 April 2018
"Characterizing and managing missing structured data in electronic health records: Data analysis"
Missing data is a challenge for all studies; however, this is especially true for electronic health record (EHR)-based analyses. Failure to appropriately consider missing data can lead to biased results. While there has been extensive theoretical work on imputation, and many sophisticated methods are now available, it remains quite challenging for researchers to implement these methods appropriately. Here, we provide detailed procedures for when and how to conduct imputation of EHR laboratory results.
The objective of this study was to demonstrate how the mechanism of "missingness" can be assessed, evaluate the performance of a variety of imputation methods, and describe some of the most frequent problems that can be encountered. (Full article...)
Recently featured: