Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 BaronePLOSCompBio2017 13-11.png|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Unmet needs for analyzing biological big data: A survey of 704 NSF principal investigators|Unmet needs for analyzing biological big data: A survey of 704 NSF principal investigators]]"''' | ||
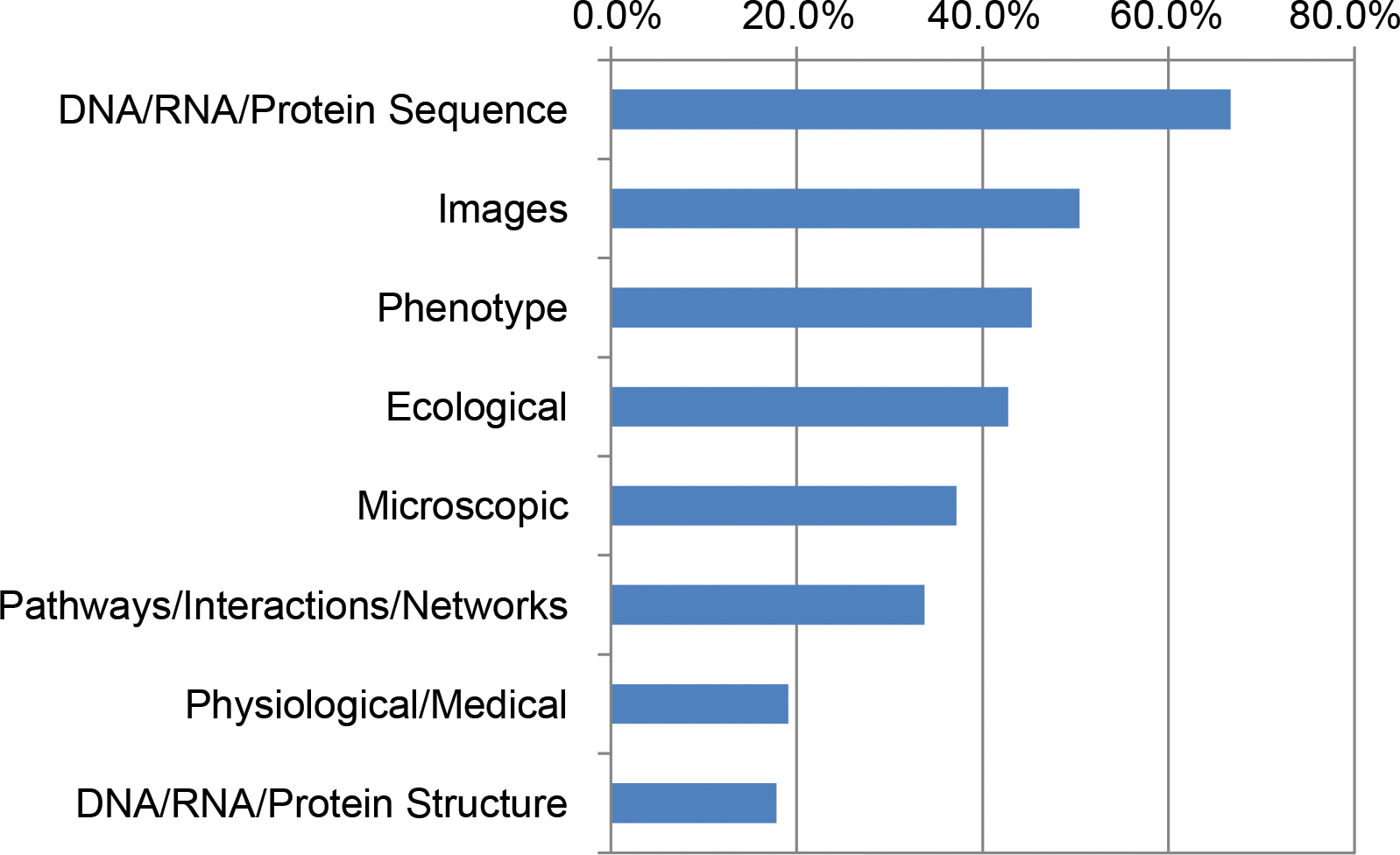
In a 2016 survey of 704 National Science Foundation (NSF) Biological Sciences Directorate principal investigators (BIO PIs), nearly 90% indicated they are currently or will soon be analyzing large data sets. BIO PIs considered a range of computational needs important to their work, including high-performance computing (HPC), [[bioinformatics]] support, multistep workflows, updated analysis software, and the ability to store, share, and publish data. Previous studies in the United States and Canada emphasized infrastructure needs. However, BIO PIs said the most pressing unmet needs are training in data integration, data management, and scaling analyses for HPC, acknowledging that data science skills will be required to build a deeper understanding of life. This portends a growing data knowledge gap in biology and challenges institutions and funding agencies to redouble their support for computational training in biology. ('''[[Journal:Unmet needs for analyzing biological big data: A survey of 704 NSF principal investigators|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Big data as a driver for clinical decision support systems: A learning health systems perspective|Big data as a driver for clinical decision support systems: A learning health systems perspective]] | |||
: ▪ [[Journal:Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory|Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory]] | : ▪ [[Journal:Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory|Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory]] | ||
: ▪ [[Journal:An open experimental database for exploring inorganic materials|An open experimental database for exploring inorganic materials]] | : ▪ [[Journal:An open experimental database for exploring inorganic materials|An open experimental database for exploring inorganic materials]] | ||
Revision as of 14:15, 26 June 2018
"Unmet needs for analyzing biological big data: A survey of 704 NSF principal investigators"
In a 2016 survey of 704 National Science Foundation (NSF) Biological Sciences Directorate principal investigators (BIO PIs), nearly 90% indicated they are currently or will soon be analyzing large data sets. BIO PIs considered a range of computational needs important to their work, including high-performance computing (HPC), bioinformatics support, multistep workflows, updated analysis software, and the ability to store, share, and publish data. Previous studies in the United States and Canada emphasized infrastructure needs. However, BIO PIs said the most pressing unmet needs are training in data integration, data management, and scaling analyses for HPC, acknowledging that data science skills will be required to build a deeper understanding of life. This portends a growing data knowledge gap in biology and challenges institutions and funding agencies to redouble their support for computational training in biology. (Full article...)
Recently featured: