Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig2 Backman BMCBio2016 17.gif|240px]]</div> | ||
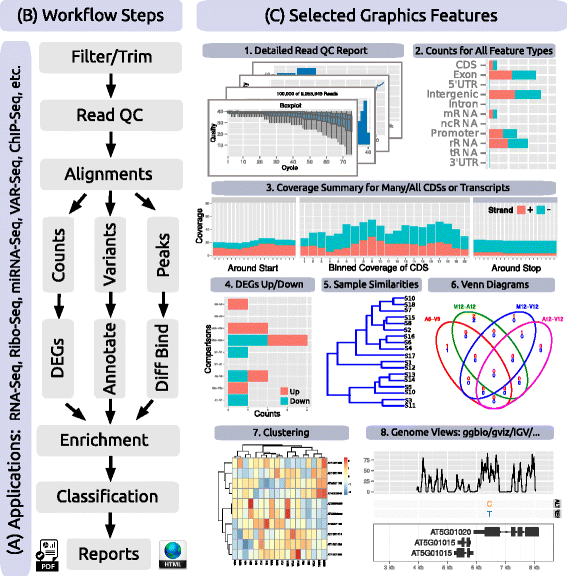
'''"[[Journal: | '''"[[Journal:systemPipeR: NGS workflow and report generation environment|systemPipeR: NGS workflow and report generation environment]]"''' | ||
Next-generation sequencing (NGS) has revolutionized how research is carried out in many areas of biology and medicine. However, the analysis of NGS data remains a major obstacle to the efficient utilization of the technology, as it requires complex multi-step processing of big data, demanding considerable computational expertise from users. While substantial effort has been invested on the development of software dedicated to the individual analysis steps of NGS experiments, insufficient resources are currently available for integrating the individual software components within the widely used R/Bioconductor environment into automated [[Workflow|workflows]] capable of running the analysis of most types of NGS applications from start-to-finish in a time-efficient and reproducible manner. ('''[[Journal:systemPipeR: NGS workflow and report generation environment|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:A data quality strategy to enable FAIR, programmatic access across large, diverse data collections for high performance data analysis|A data quality strategy to enable FAIR, programmatic access across large, diverse data collections for high performance data analysis]] | |||
: ▪ [[Journal:How big data, comparative effectiveness research, and rapid-learning health care systems can transform patient care in radiation oncology|How big data, comparative effectiveness research, and rapid-learning health care systems can transform patient care in radiation oncology]] | : ▪ [[Journal:How big data, comparative effectiveness research, and rapid-learning health care systems can transform patient care in radiation oncology|How big data, comparative effectiveness research, and rapid-learning health care systems can transform patient care in radiation oncology]] | ||
: ▪ [[Journal:Wireless positioning in IoT: A look at current and future trends|Wireless positioning in IoT: A look at current and future trends]] | : ▪ [[Journal:Wireless positioning in IoT: A look at current and future trends|Wireless positioning in IoT: A look at current and future trends]] | ||
Revision as of 15:02, 24 October 2018
"systemPipeR: NGS workflow and report generation environment"
Next-generation sequencing (NGS) has revolutionized how research is carried out in many areas of biology and medicine. However, the analysis of NGS data remains a major obstacle to the efficient utilization of the technology, as it requires complex multi-step processing of big data, demanding considerable computational expertise from users. While substantial effort has been invested on the development of software dedicated to the individual analysis steps of NGS experiments, insufficient resources are currently available for integrating the individual software components within the widely used R/Bioconductor environment into automated workflows capable of running the analysis of most types of NGS applications from start-to-finish in a time-efficient and reproducible manner. (Full article...)
Recently featured:
- ▪ A data quality strategy to enable FAIR, programmatic access across large, diverse data collections for high performance data analysis
- ▪ How big data, comparative effectiveness research, and rapid-learning health care systems can transform patient care in radiation oncology
- ▪ Wireless positioning in IoT: A look at current and future trends