Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text) |
Shawndouglas (talk | contribs) (Updated article of the week text) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Bhattacharya FrontInOnc2019 9.jpg|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:AI meets exascale computing: Advancing cancer research with large-scale high-performance computing|AI meets exascale computing: Advancing cancer research with large-scale high-performance computing]]"''' | ||
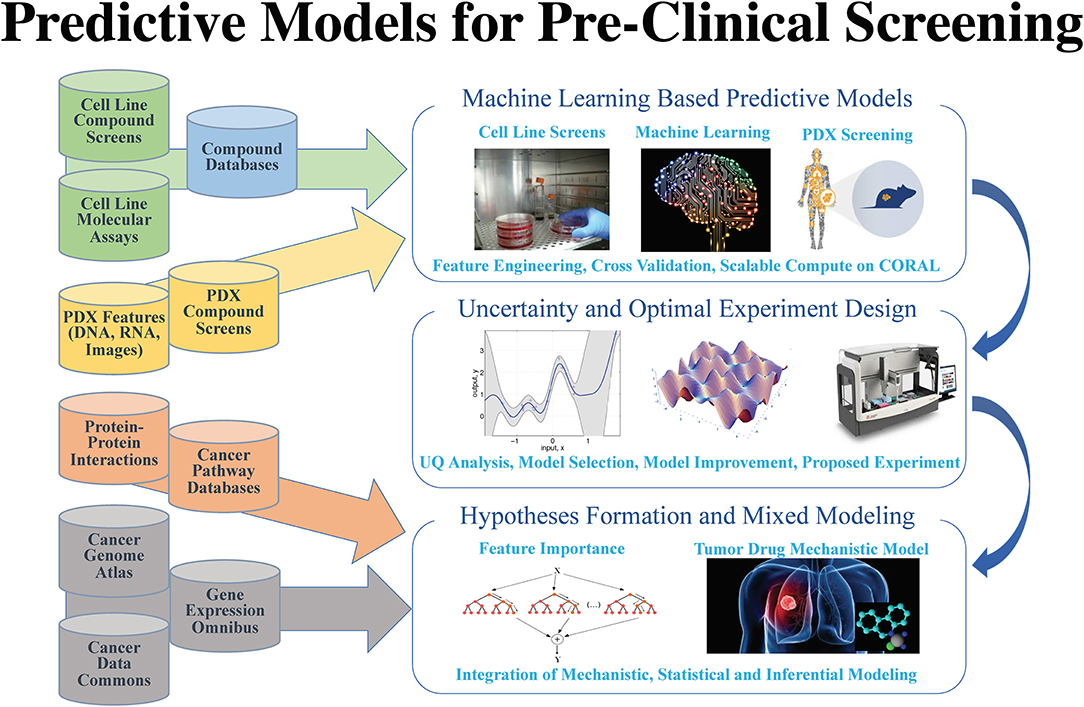
The application of data science in [[cancer]] research has been boosted by major advances in three primary areas: (1) data: diversity, amount, and availability of biomedical data; (2) advances in [[artificial intelligence]] (AI) and machine learning (ML) algorithms that enable learning from complex, large-scale data; and (3) advances in computer architectures allowing unprecedented acceleration of simulation and machine learning algorithms. These advances help build ''in silico'' ML models that can provide transformative insights from data, including molecular dynamics simulations, [[Sequencing|next-generation sequencing]], omics, [[Molecular imaging|imaging]], and unstructured clinical text documents. Unique challenges persist, however, in building ML models related to cancer, including: (1) access, sharing, labeling, and integration of multimodal and multi-institutional data across different cancer types; (2) developing AI models for cancer research capable of scaling on next-generation high-performance computers; and (3) assessing robustness and reliability in the AI models. ('''[[Journal:AI meets exascale computing: Advancing cancer research with large-scale high-performance computing|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Building infrastructure for African human genomic data management|Building infrastructure for African human genomic data management]] | |||
: ▪ [[Journal:Process variation detection using missing data in a multihospital community practice anatomic pathology laboratory|Process variation detection using missing data in a multihospital community practice anatomic pathology laboratory]] | : ▪ [[Journal:Process variation detection using missing data in a multihospital community practice anatomic pathology laboratory|Process variation detection using missing data in a multihospital community practice anatomic pathology laboratory]] | ||
: ▪ [[Journal:Development and validation of a fast gas chromatography–mass spectrometry method for the determination of cannabinoids in Cannabis sativa L|Development and validation of a fast gas chromatography–mass spectrometry method for the determination of cannabinoids in Cannabis sativa L]] | : ▪ [[Journal:Development and validation of a fast gas chromatography–mass spectrometry method for the determination of cannabinoids in Cannabis sativa L|Development and validation of a fast gas chromatography–mass spectrometry method for the determination of cannabinoids in Cannabis sativa L]] | ||
Revision as of 15:47, 8 January 2020
"AI meets exascale computing: Advancing cancer research with large-scale high-performance computing"
The application of data science in cancer research has been boosted by major advances in three primary areas: (1) data: diversity, amount, and availability of biomedical data; (2) advances in artificial intelligence (AI) and machine learning (ML) algorithms that enable learning from complex, large-scale data; and (3) advances in computer architectures allowing unprecedented acceleration of simulation and machine learning algorithms. These advances help build in silico ML models that can provide transformative insights from data, including molecular dynamics simulations, next-generation sequencing, omics, imaging, and unstructured clinical text documents. Unique challenges persist, however, in building ML models related to cancer, including: (1) access, sharing, labeling, and integration of multimodal and multi-institutional data across different cancer types; (2) developing AI models for cancer research capable of scaling on next-generation high-performance computers; and (3) assessing robustness and reliability in the AI models. (Full article...)
Recently featured:
- ▪ Building infrastructure for African human genomic data management
- ▪ Process variation detection using missing data in a multihospital community practice anatomic pathology laboratory
- ▪ Development and validation of a fast gas chromatography–mass spectrometry method for the determination of cannabinoids in Cannabis sativa L