Difference between revisions of "User:Shawndouglas/sandbox/sublevel9"
Shawndouglas (talk | contribs) |
Shawndouglas (talk | contribs) |
||
| Line 108: | Line 108: | ||
|} | |} | ||
|} | |} | ||
==Use cases== | |||
To better illustrate the use of the competencies, we present here a series of brief use cases—scenarios in which the competencies have proven valuable already in defining, refining, or assessing a bioinformatics training mechanism. These use cases were selected to highlight a diverse set of training needs, user personas, types of training programs, and educational settings. In this spirit, we present examples grouped into three categories: (1) complete degree programs for which the competencies have proven valuable to overall curriculum design or refinement; (2) supplements to existing degree programs (i.e., specializations, tracks, certificates); and (3) training resources outside the context of specific degree programs. | |||
===Degree programs=== | |||
====Introductory and masters bioinformatics training in Africa: H3ABioNet==== | |||
H3ABioNet (www.h3abionet.org), a Pan African bioinformatics network for H3Africa<ref name="MulderH3ABioNet16">{{cite journal |title=H3ABioNet, a sustainable pan-African bioinformatics network for human heredity and health in Africa |journal=Genome Research |author=Mulder, N.J.; Adebiyi, E.; Alami, R. et al. |volume=26 |issue=2 |pages=271-7 |year=2016 |doi=10.1101/gr.196295.115 |pmid=26627985 |pmc=PMC4728379}}</ref>, has developed a bioinformatics training program for African scientists from the Human Heredity and Health in Africa (www.h3africa.org) consortium. This involves bioinformatics training for a broad range of audiences, primarily in [[genomics]] [[data analysis]], and the development of new bioinformatics degrees to train bioinformatics scientists. Though there are some institutions in Africa offering bioinformatics postgraduate degrees, this was limited to a handful of countries, and many additional institutions expressed a desire to develop and offer such degrees in order to build the next generation of bioinformatics academics. An African Bioinformatics Education Committee was established along with a Curriculum Task Force, which set about designing a bioinformatics master’s program. Topic areas were selected from existing master’s courses and those proposed by Welch ''et al.''.<ref name="WelchBioinformatics14" /> From these, core modules were defined and augmented with additional elective modules relevant to specific institutions, based on their research priorities. The Curriculum Task Force then fleshed out the detailed content of each module and started mapping these to core competencies required of a bioinformatics specialist. Though the focus of some master’s programs may vary from the more biological to a stronger emphasis on software engineering, there were common competencies with which all bioinformatics master’s graduates should be equipped. While some African institutions have specific research focus areas, the feeling was that all students training to be bioinformaticians should be exposed to a set of core subjects, which are in line with the ISCB’s recommendations, and the elective subjects then tend to be dependent on the research focus. The proposed curriculum has been put into practice, with at least two universities in Africa starting their first master’s programs in the last two years. | |||
For bioinformatics users, H3ABioNet has successfully run several specialist short courses to train researchers on next generation sequence analysis, metagenomics, genome-wide association studies, and other topics. However, through interactions with users, there emerged a need for more basic “introduction to bioinformatics” training. In response, H3ABioNet developed an Introduction to Bioinformatics course delivered remotely to classrooms across multiple countries. The curriculum was derived primarily from topics used for the master’s courses, but this time mapping it to competencies for bioinformatics users and removing topics with a modelling or programming focus. The competencies for this audience are thus more focused on a basic understanding of the topic, example algorithms, and how the tools can be applied to answer biological questions. The practicals are also designed to enable users to navigate their way through the tools and learn to interpret the outputs. This course was run successfully for the first time in 2016 and was assessed to determine whether the required core competencies were acquired. | |||
Using core competencies for both cases described above enabled course organizers to better define the detailed content, contact hours, and focus for each module, based on the intended audience. We could also use the competencies to define learning outcomes and refine module assessments. | |||
====Undergraduate and graduate degree programs in a U.S. research university: Computational biology education at Carnegie Mellon==== | |||
Carnegie Mellon University has long been active in education in computational biology and bioinformatics, providing several opportunities for considering how a general set of competencies can apply to diverse populations. These experiences include degree programs in computational biology at several levels, including a bachelor's of science (BS) in computational biology (since 1989), a masters of science (MS) in computational biology (since 1999), a doctorate (PhD) in computational biology (offered jointly with the University of Pittsburgh since 2005), and required training in computational biology as part of the core of the BS in biological sciences, the university's general undergraduate biology major. While all of these programs predate the ISCB competencies, the competencies provide a basis for considering how well these programs prepare students for work involving computational biology to differing degrees. Two of these programs—the BS in biological sciences and the PhD in computational biology—are discussed as examples of programs with very different student populations and training needs that can be evaluated in light of the competencies. | |||
Carnegie Mellon's BS in biological sciences illustrates one kind of bioinformatics training: for students primarily training for work in experimental biology. Carnegie Mellon took the still unusual step in 2013 of making Introduction to Computational Biology (ICB) a core requirement of every undergraduate biological sciences major, providing an opportunity to explore how one would design a class to be accessible but rigorous and useful to a population of general biology students. Applying these competencies, then, requires working in the context of students who are typically taking a single class on computational biology but within a full undergraduate biology curriculum. Some competencies, primarily those focused on technical aspects of computational biology, can be covered reasonably well at the level needed by an experimental biologist within a single computational biology class (C,D,F,I,J; see Table 1). Other important areas, such as more conventional biological knowledge, are covered thoroughly in other areas of an undergraduate biological sciences curriculum, e.g., in more traditional core classes such as genetics, biochemistry, or cell biology (A,B). Still others, such as the topics that fall broadly under communications and professional development, are covered elsewhere in the curriculum by a variety of mechanisms inside and outside the classroom (M,N,O,P). Still other areas go beyond what can fit in one introductory class but are also not covered elsewhere. Some of these (G,H,K) are competencies that may not be needed by this population but can be flagged for consideration in revisions of ICB. The most interesting topics are those that are crucial for experimental biologists, cannot be covered sufficiently in ICB, and are not covered elsewhere (E, i.e., biostatistics). ICB gives this latter area enough coverage to convey the key ideas needed for bioinformatics work, but the competencies flag it as an area in need of further development in the curriculum as a whole. | |||
The Carnegie Mellon/University of Pittsburgh joint PhD in computational biology offers an example at another extreme of the spectrum: a full multi-year training program for students expected to become experts in computational biology, who are expected to graduate competent to lead independent research programs in the area, teach computational biology, run bioinformatics core facilities, or pursue similarly demanding jobs. Computational biology programs face a special challenge compared with more traditional degree programs, in that the lack of clear standards for training at the undergraduate level means that there is little one can assume or enforce about background knowledge of incoming students beyond basic competencies in biology, computing, and mathematics. Furthermore, since a PhD program is research-focused and under pressure to limit time to degree, formal training can occupy only a finite amount of a student's time, equivalent to roughly a year of full-time coursework. To a limited degree, the program can rely on admissions standards, remediation, and self-teaching to assume some basics of all students (A,F,I,J). Some competencies can be handled by flexible menu-based requirements to meet a competency in ways appropriate to each student’s individual needs and background (B). In others, every student needs a high level of competency, and this must be met with specialized core classes designed for this population (C,D,E,G,H). Others must be met within the curriculum through specialized professional development mechanisms as well as one-on-one mentorship by the thesis advisor (K,L,M,N,O,P). Nonetheless, some competencies, especially those that depend on the mentorship of the research advisor, may be acquired much more effectively by some students than others. The competencies again suggest that these topics should be flagged for consideration for more formal training in the future. Furthermore, the challenges faced by this program with respect to knowledge of incoming students make clear the value that accepted standards for competencies at the undergraduate level could have in making most effective use of time in graduate school for specialists in the field. | |||
==References== | ==References== | ||
Revision as of 17:41, 26 August 2018
|
|
This is sublevel2 of my sandbox, where I play with features and test MediaWiki code. If you wish to leave a comment for me, please see my discussion page instead. |
Sandbox begins below
| Full article title | The development and application of bioinformatics core competencies to improve bioinformatics training and education |
|---|---|
| Journal | PLOS Computational Biology |
| Author(s) |
Mulder, Nicola; Schwartz, Russell; Brazas, Michelle, D.; Brooksbank, Carth; Gaeta, Bruno; Morgan, Sarah L.; Pauley, Mark A.; Rosenwald, Anne; Rustici, Gabriella; Sierk, Michael; Warnow, Tandy; Welch, Lonnie |
| Author affiliation(s) |
University of Cape Town, Carnegie Mellon University, Ontario Institute for Cancer Research, Wellcome Genome Campus, University of New South Wales, University of Nebraska at Omaha, Georgetown University, University of Cambridge, Saint Vincent College, University of Illinois at Urbana-Champaign, Ohio University |
| Primary contact | Email: nicola dot mulder at uct dot ac dot za |
| Year published | 2018 |
| Volume and issue | 14(2) |
| Page(s) | e1005772 |
| DOI | 10.1371/journal.pcbi.1005772 |
| ISSN | 1553-7358 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005772 |
| Download | http://journals.plos.org/ploscompbiol/article/file?id=10.1371/journal.pcbi.1005772&type=printable (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Bioinformatics is recognized as part of the essential knowledge base of numerous career paths in biomedical research and healthcare. However, there is little agreement in the field over what that knowledge entails or how best to provide it. These disagreements are compounded by the wide range of populations in need of bioinformatics training, with divergent prior backgrounds and intended application areas. The Curriculum Task Force of the International Society of Computational Biology (ISCB) Education Committee has sought to provide a framework for training needs and curricula in terms of a set of bioinformatics core competencies that cut across many user personas and training programs. The initial competencies developed based on surveys of employers and training programs have since been refined through a multiyear process of community engagement. This report describes the current status of the competencies and presents a series of use cases illustrating how they are being applied in diverse training contexts. These use cases are intended to demonstrate how others can make use of the competencies and engage in the process of their continuing refinement and application. The report concludes with a consideration of remaining challenges and future plans.
Introduction
The need for bioinformatics education and training is immense, but it is also diverse. There is a wide range of audiences who are potential recipients of training, each of which has different needs in terms of what skills or knowledge they require and at what depth. For example, someone training to be a bioinformatics engineer (which we define as someone who will actively be involved in the development and application of bioinformatics algorithms) requires in-depth knowledge of existing algorithms, how they work, how to critically evaluate them, and how to interpret the results. By contrast, a bioinformatics user (which we define as someone making use of bioinformatics resources in an applied context, such as in medical practice) would need a basic level of understanding of the methods and a stronger focus on the interpretation of the outputs. In a recent publication[1], the ISCB Education Committee’s Curriculum Task Force described the potential for refinement and application of bioinformatics core competencies for different user groups. Here, we describe the further refinement of these competencies and provide a series of use cases illustrating their applications to different bioinformatics education and training programs globally.
Development of core competencies for bioinformatics
The ISCB Curriculum Task Force undertook the task of identifying some of the breadth of needs for bioinformatics education, as described in a series of reports from the task force. This effort arose first from a series of surveys of current training practice and desired training needs[2], which identified a set of broad categories of training needs but also widespread disparities across programs in what was taught, how, and for what intended target audiences. An outcome of these surveys was the need for identifying a set of core competencies as broad categories of skills and training that cross different programs and training needs and that can provide a basis for discussing similarities and differences between programs and desired outcomes. This led to a further effort to define a set of initial core competencies[3] that in turn led to an intensive program of community engagement to refine these competencies to better serve the breadth of needs of the bioinformatics training community.
There were three major steps in the development of the core competencies: (1) defining the competencies needed for using bioinformatics, (2) defining a variety of user profiles describing distinct subgroups in need of training, and (3) defining how the competencies will apply to each user profile (scoring). The core competency framework was developed through an iterative process with input from multiple parties from diverse backgrounds with a connection to bioinformatics. In order to gain a broader appreciation of which competencies the bioinformatics community considers relevant for different bioinformatics user profiles, the ISCB Curriculum Task Force has run several competency workshops (discussion sessions for defining the competencies and their applications) both at ISCB conferences and at other bioinformatics education venues such as the GOBLET (Global Organisation for Bioinformatics Learning, Education and Training) Annual General Meeting. Each iteration of a competency workshop has greatly enhanced not only the competencies themselves but also the definitions of the user profiles[1] and the competency-use case scoring mechanism.
Initially, the mapping of bioinformatics competencies to audiences considered three major user profiles: (1) the bioinformatics user; (2) the bioinformatics scientist; and (3) the bioinformatics engineer. Early competency workshops quickly surmised that these user profiles were too narrow and did not adequately capture the breadth of roles requiring bioinformatics competency and curriculum. Participants spent much of the workshop time defining a bioinformatics user or distinguishing a bioinformatics scientist from a bioinformatics engineer. The use case roles were subsequently expanded to better embody the breadth of bioinformatics users, including physicians, lab technicians, ethicists and biocurators, scientists (which include the discovery biologist, academic bioinformatics researcher, and core facility scientist), and engineers (which may be a bioinformatician in academia, bioinformatician in research institute, or software engineer). This change allowed for subsequent workshop participants to self-select according to the category of user with which they most identified.
With user profiles better defined, competency workshops then struggled with the competencies themselves and their definitions. Several early competency definitions appeared to overlap. For example, “Apply knowledge of computing appropriate to the discipline (e.g., effectively utilize bioinformatics tools)” closely resembled “Analyze a problem and identify and define the computing requirements appropriate to its solution (e.g., define algorithmic time and space complexities and hardware resources required to solve a problem).” Workshop participants helped to reduce the redundancy in our initial set of bioinformatics competencies from 20 competencies to a refined set of 16 competencies.
Competency workshops have additionally helped to revise the scoring of competencies for each user profile. Early workshops scored the applicability of a bioinformatics competency to a particular profile with a simple yes/no response, which did not allow for an appreciation of the depth of the competency necessary for a given profile. Such a scoring approach, while better than no score, would not be helpful when developing a curriculum for a specific user profile. Subsequent workshops used a graded scoring approach, with grades ranging from 1 (no competency required) to 4 (specialist knowledge required). This, too, proved too ambiguous to allow for meaningful discussion and classification. The scoring approach was thus revised again to the current model, which uses the Bloom’s Revised Taxonomy[4] terms: knowledge, comprehension, application, analysis, synthesis, and evaluation. While the use of Bloom’s Taxonomy has been useful in mapping competency levels to each of the user profiles, this change required refinement of the competency list as several of the earlier competencies incorporated Bloom’s Taxonomy terms.
Overall, competency workshops have been invaluable to the enhancement and refinement of the bioinformatics competencies. Through these workshops, the ISCB Curriculum Task Force has been able to construct a useful set of bioinformatics competencies that curriculum developers can use to develop, compare, and assess impactful bioinformatics training programs for a wide range of audiences and ultimately help establish bioinformatics skills in such audiences.[3]
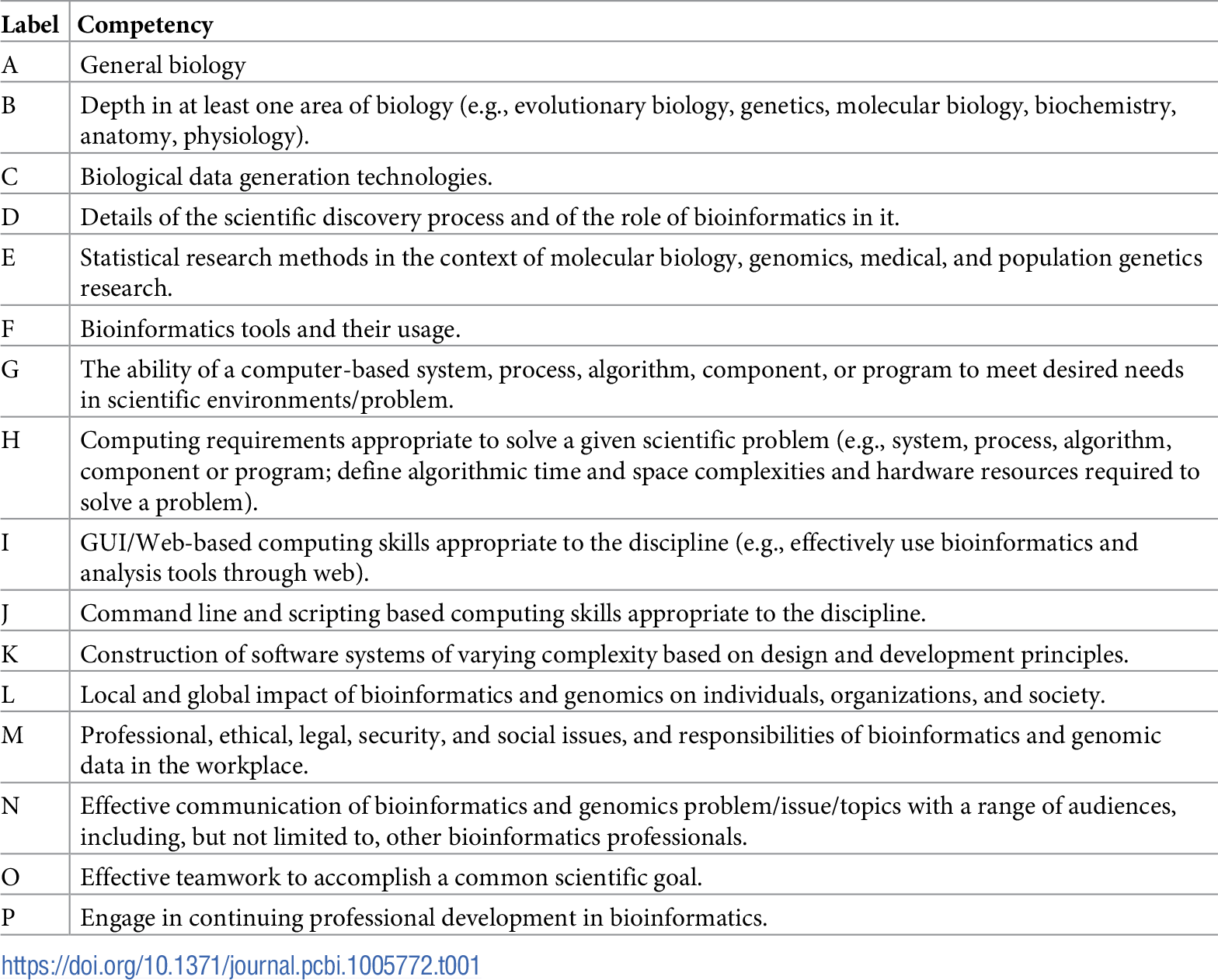
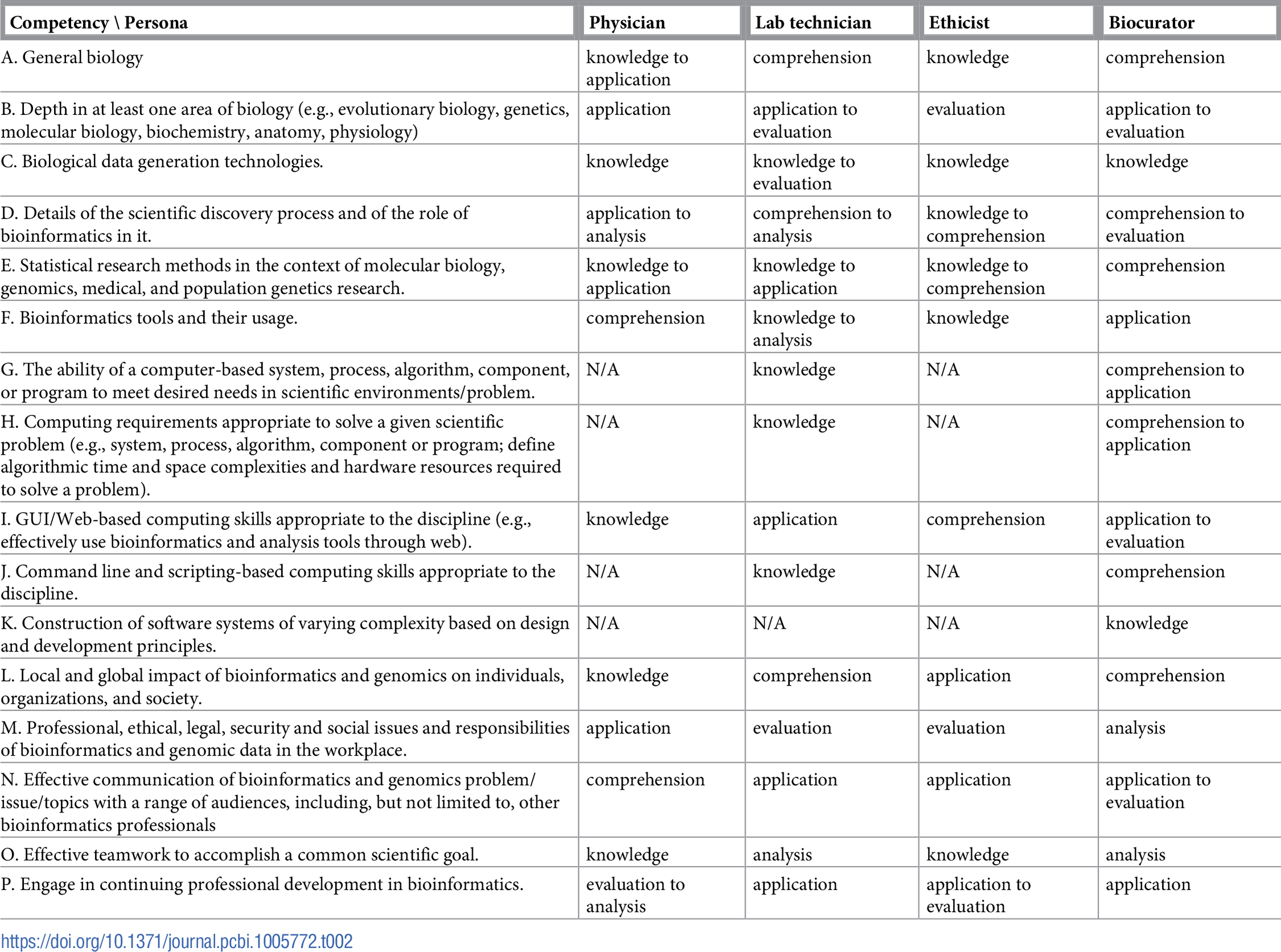
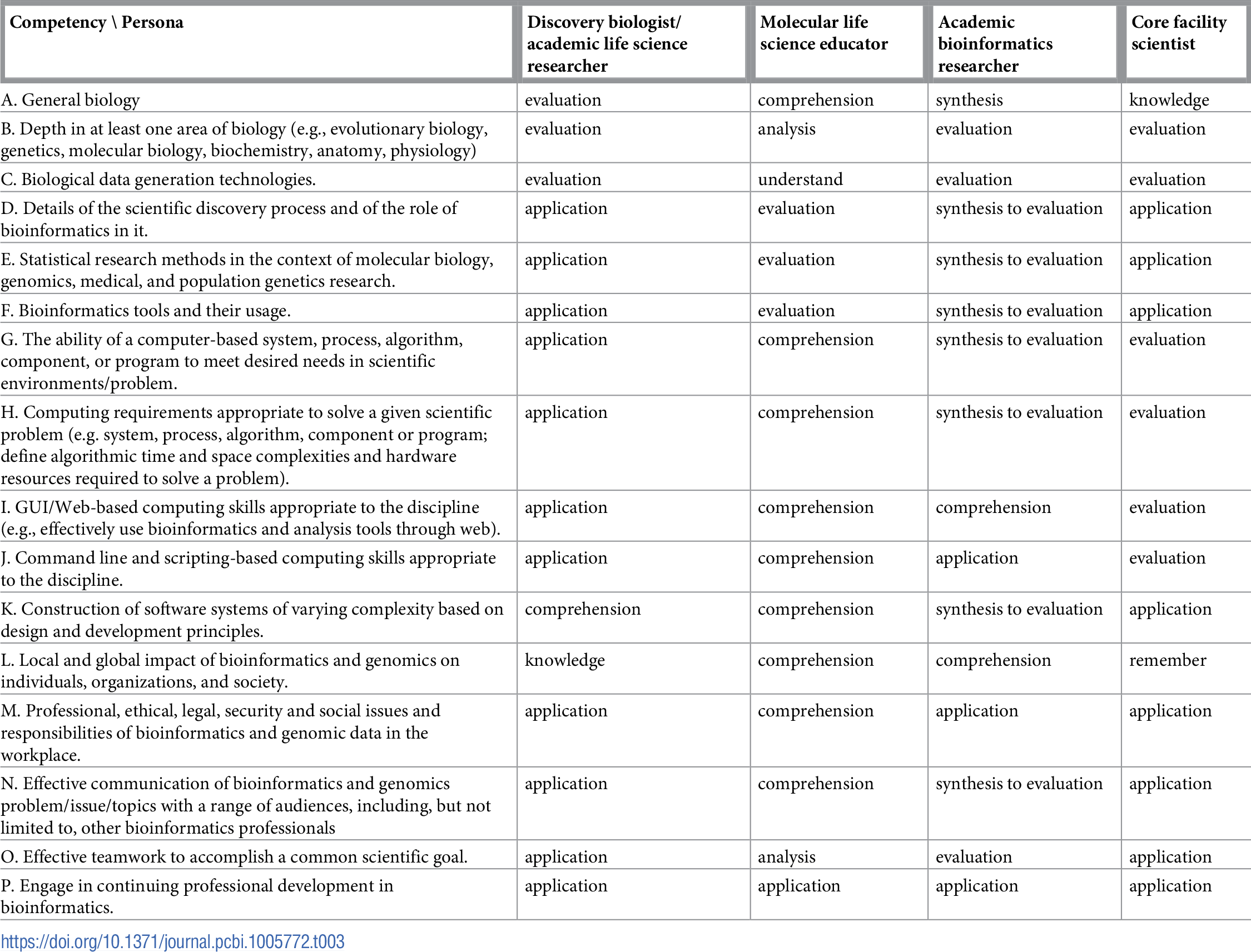
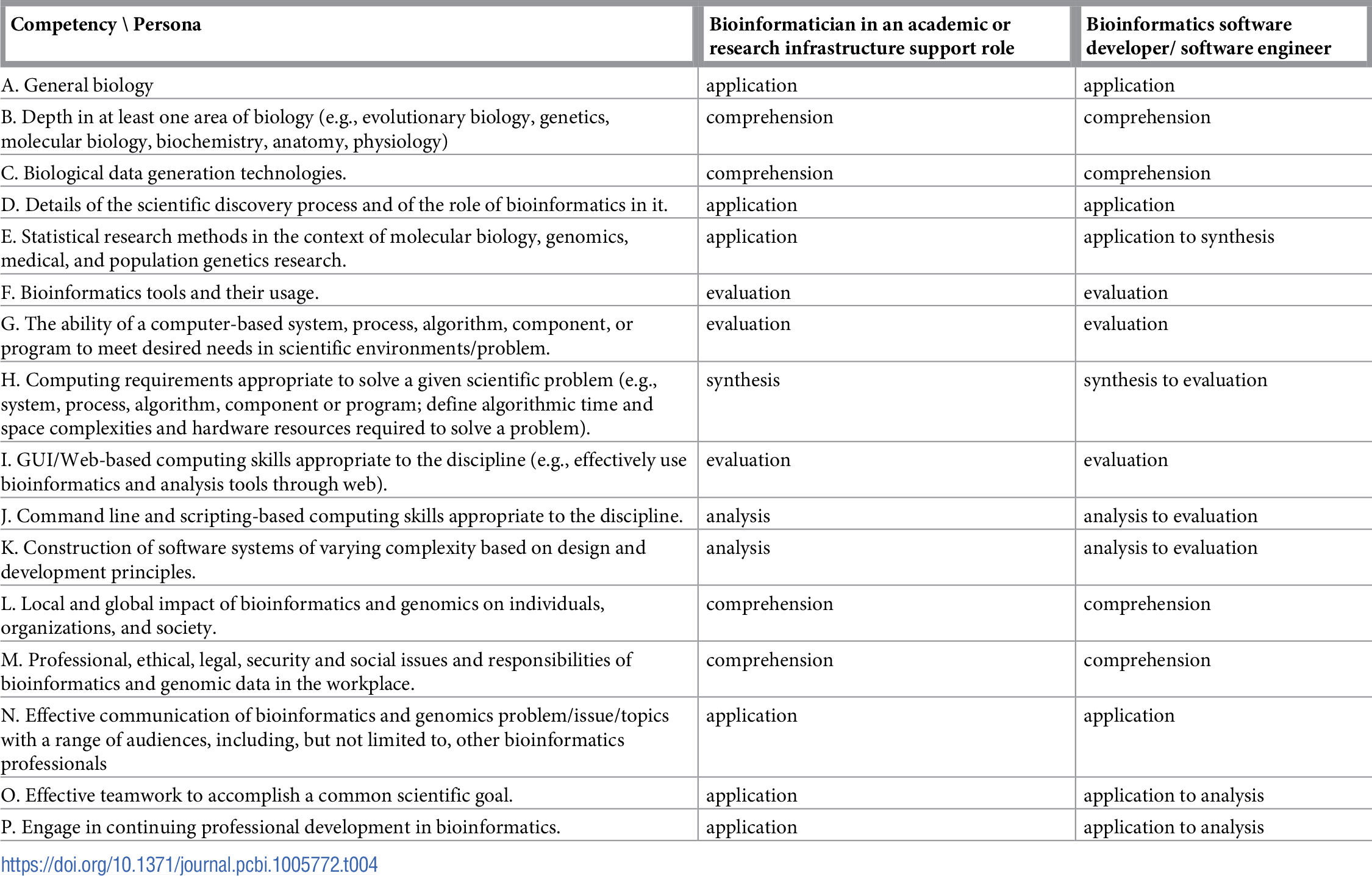
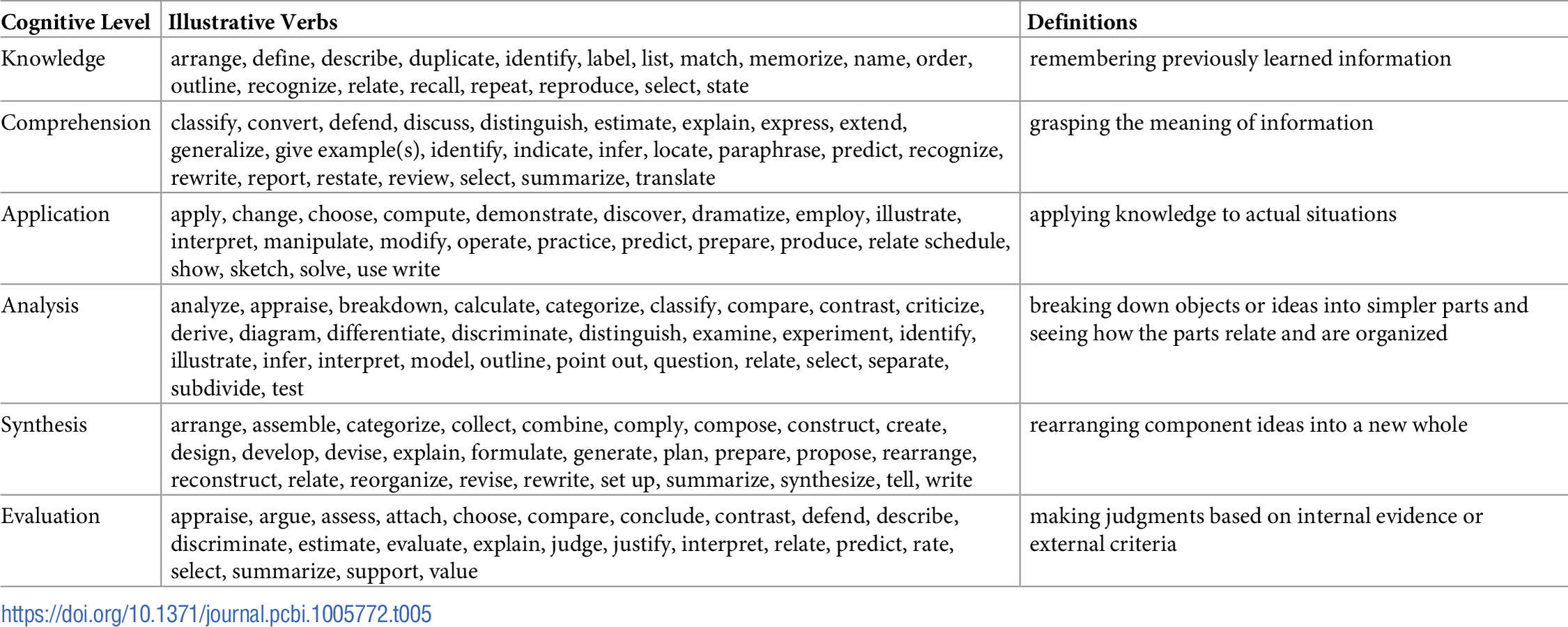
Table 1 reports the current state of the competencies developed and refined through this community engagement process. Tables 2–4 map these refined competencies to a broader set of personas, suggested over the course of the Task Force’s community engagement efforts, via Bloom’s Taxonomy terms. For reference, Table 5 provides examples and definitions of the Bloom's Revised Taxonomy terms. In the next section, we provide some examples of how the competencies have been applied in a variety of training contexts.
|
|
|
|
|
Use cases
To better illustrate the use of the competencies, we present here a series of brief use cases—scenarios in which the competencies have proven valuable already in defining, refining, or assessing a bioinformatics training mechanism. These use cases were selected to highlight a diverse set of training needs, user personas, types of training programs, and educational settings. In this spirit, we present examples grouped into three categories: (1) complete degree programs for which the competencies have proven valuable to overall curriculum design or refinement; (2) supplements to existing degree programs (i.e., specializations, tracks, certificates); and (3) training resources outside the context of specific degree programs.
Degree programs
Introductory and masters bioinformatics training in Africa: H3ABioNet
H3ABioNet (www.h3abionet.org), a Pan African bioinformatics network for H3Africa[5], has developed a bioinformatics training program for African scientists from the Human Heredity and Health in Africa (www.h3africa.org) consortium. This involves bioinformatics training for a broad range of audiences, primarily in genomics data analysis, and the development of new bioinformatics degrees to train bioinformatics scientists. Though there are some institutions in Africa offering bioinformatics postgraduate degrees, this was limited to a handful of countries, and many additional institutions expressed a desire to develop and offer such degrees in order to build the next generation of bioinformatics academics. An African Bioinformatics Education Committee was established along with a Curriculum Task Force, which set about designing a bioinformatics master’s program. Topic areas were selected from existing master’s courses and those proposed by Welch et al..[3] From these, core modules were defined and augmented with additional elective modules relevant to specific institutions, based on their research priorities. The Curriculum Task Force then fleshed out the detailed content of each module and started mapping these to core competencies required of a bioinformatics specialist. Though the focus of some master’s programs may vary from the more biological to a stronger emphasis on software engineering, there were common competencies with which all bioinformatics master’s graduates should be equipped. While some African institutions have specific research focus areas, the feeling was that all students training to be bioinformaticians should be exposed to a set of core subjects, which are in line with the ISCB’s recommendations, and the elective subjects then tend to be dependent on the research focus. The proposed curriculum has been put into practice, with at least two universities in Africa starting their first master’s programs in the last two years.
For bioinformatics users, H3ABioNet has successfully run several specialist short courses to train researchers on next generation sequence analysis, metagenomics, genome-wide association studies, and other topics. However, through interactions with users, there emerged a need for more basic “introduction to bioinformatics” training. In response, H3ABioNet developed an Introduction to Bioinformatics course delivered remotely to classrooms across multiple countries. The curriculum was derived primarily from topics used for the master’s courses, but this time mapping it to competencies for bioinformatics users and removing topics with a modelling or programming focus. The competencies for this audience are thus more focused on a basic understanding of the topic, example algorithms, and how the tools can be applied to answer biological questions. The practicals are also designed to enable users to navigate their way through the tools and learn to interpret the outputs. This course was run successfully for the first time in 2016 and was assessed to determine whether the required core competencies were acquired.
Using core competencies for both cases described above enabled course organizers to better define the detailed content, contact hours, and focus for each module, based on the intended audience. We could also use the competencies to define learning outcomes and refine module assessments.
Undergraduate and graduate degree programs in a U.S. research university: Computational biology education at Carnegie Mellon
Carnegie Mellon University has long been active in education in computational biology and bioinformatics, providing several opportunities for considering how a general set of competencies can apply to diverse populations. These experiences include degree programs in computational biology at several levels, including a bachelor's of science (BS) in computational biology (since 1989), a masters of science (MS) in computational biology (since 1999), a doctorate (PhD) in computational biology (offered jointly with the University of Pittsburgh since 2005), and required training in computational biology as part of the core of the BS in biological sciences, the university's general undergraduate biology major. While all of these programs predate the ISCB competencies, the competencies provide a basis for considering how well these programs prepare students for work involving computational biology to differing degrees. Two of these programs—the BS in biological sciences and the PhD in computational biology—are discussed as examples of programs with very different student populations and training needs that can be evaluated in light of the competencies.
Carnegie Mellon's BS in biological sciences illustrates one kind of bioinformatics training: for students primarily training for work in experimental biology. Carnegie Mellon took the still unusual step in 2013 of making Introduction to Computational Biology (ICB) a core requirement of every undergraduate biological sciences major, providing an opportunity to explore how one would design a class to be accessible but rigorous and useful to a population of general biology students. Applying these competencies, then, requires working in the context of students who are typically taking a single class on computational biology but within a full undergraduate biology curriculum. Some competencies, primarily those focused on technical aspects of computational biology, can be covered reasonably well at the level needed by an experimental biologist within a single computational biology class (C,D,F,I,J; see Table 1). Other important areas, such as more conventional biological knowledge, are covered thoroughly in other areas of an undergraduate biological sciences curriculum, e.g., in more traditional core classes such as genetics, biochemistry, or cell biology (A,B). Still others, such as the topics that fall broadly under communications and professional development, are covered elsewhere in the curriculum by a variety of mechanisms inside and outside the classroom (M,N,O,P). Still other areas go beyond what can fit in one introductory class but are also not covered elsewhere. Some of these (G,H,K) are competencies that may not be needed by this population but can be flagged for consideration in revisions of ICB. The most interesting topics are those that are crucial for experimental biologists, cannot be covered sufficiently in ICB, and are not covered elsewhere (E, i.e., biostatistics). ICB gives this latter area enough coverage to convey the key ideas needed for bioinformatics work, but the competencies flag it as an area in need of further development in the curriculum as a whole.
The Carnegie Mellon/University of Pittsburgh joint PhD in computational biology offers an example at another extreme of the spectrum: a full multi-year training program for students expected to become experts in computational biology, who are expected to graduate competent to lead independent research programs in the area, teach computational biology, run bioinformatics core facilities, or pursue similarly demanding jobs. Computational biology programs face a special challenge compared with more traditional degree programs, in that the lack of clear standards for training at the undergraduate level means that there is little one can assume or enforce about background knowledge of incoming students beyond basic competencies in biology, computing, and mathematics. Furthermore, since a PhD program is research-focused and under pressure to limit time to degree, formal training can occupy only a finite amount of a student's time, equivalent to roughly a year of full-time coursework. To a limited degree, the program can rely on admissions standards, remediation, and self-teaching to assume some basics of all students (A,F,I,J). Some competencies can be handled by flexible menu-based requirements to meet a competency in ways appropriate to each student’s individual needs and background (B). In others, every student needs a high level of competency, and this must be met with specialized core classes designed for this population (C,D,E,G,H). Others must be met within the curriculum through specialized professional development mechanisms as well as one-on-one mentorship by the thesis advisor (K,L,M,N,O,P). Nonetheless, some competencies, especially those that depend on the mentorship of the research advisor, may be acquired much more effectively by some students than others. The competencies again suggest that these topics should be flagged for consideration for more formal training in the future. Furthermore, the challenges faced by this program with respect to knowledge of incoming students make clear the value that accepted standards for competencies at the undergraduate level could have in making most effective use of time in graduate school for specialists in the field.
References
- ↑ 1.0 1.1 Welch, L.; Brooksban, C.; Schwartz, R. et al. (2016). "Applying, Evaluating and Refining Bioinformatics Core Competencies (An Update from the Curriculum Task Force of ISCB's Education Committee)". PLOS Computational Biology 12 (5): e1004943. doi:10.1371/journal.pcbi.1004943. PMC PMC4866758. PMID 27175996. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4866758.
- ↑ Welch, L.R.; Schwartz, R.; Lewitter, F. (2012). "A report of the Curriculum Task Force of the ISCB Education Committee". PLOS Computational Biology 8 (6): e1002570. doi:10.1371/journal.pcbi.1002570. PMC PMC3386154. PMID 22761560. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3386154.
- ↑ 3.0 3.1 3.2 Welch, L.; Lewitter, F; Schwartz, R. et al. (2014). "Bioinformatics curriculum guidelines: toward a definition of core competencies". PLOS Computational Biology 10 (3): e1003496. doi:10.1371/journal.pcbi.1003496. PMC PMC3945096. PMID 24603430. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3945096.
- ↑ Anderson, L.W.; Krathwohl, D.R., ed. (2001). A Taxonomy for Learning, Teaching, and Assessing: A Revision of Bloom's Taxonomy of Educational Objectives. Pearson. pp. 336. ISBN 9780801319037.
- ↑ Mulder, N.J.; Adebiyi, E.; Alami, R. et al. (2016). "H3ABioNet, a sustainable pan-African bioinformatics network for human heredity and health in Africa". Genome Research 26 (2): 271-7. doi:10.1101/gr.196295.115. PMC PMC4728379. PMID 26627985. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4728379.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, spelling, and grammar. PMCID and DOI were added when they were missing from the original reference.