Difference between revisions of "Journal:Infrastructure tools to support an effective radiation oncology learning health system"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 104: | Line 104: | ||
One of the major challenges with examining patients’ DICOM-RT data is the lack of standardized organs at risk (OAR) and target names, as well as ambiguity regarding dose-volume histogram metrics, and multiple prescriptions mentioned across several treatment techniques. With the goal of overcoming these challenges, the AAPM TG 263 initiative has published their recommendations on OAR and target nomenclature. The ETL user interface deploys this standardized nomenclature and requires the importer of the data to match the deemed OARs with their corresponding standard OAR and target names. In addition, this program also suggests a matching name based on an automated process of relabeling using our published techniques (OAR labels [19], radiomics features [20], and geometric information [21]). We find that these automated approaches provide an acceptable accuracy over the standard prostate and lung structure types. In order to gather the dose volume histogram data from the DICOM-RT dose and structure set files, we have deployed a DICOM-RT dosimetry parser software. If the DICOM-RT dose file exported by the TPS contains dose-volume histogram (DVH) information, we utilize it. However, if the file lacks this information, we employ our dosimetry parser software to calculate the DVH values from the dose and structure set volume information. | One of the major challenges with examining patients’ DICOM-RT data is the lack of standardized organs at risk (OAR) and target names, as well as ambiguity regarding dose-volume histogram metrics, and multiple prescriptions mentioned across several treatment techniques. With the goal of overcoming these challenges, the AAPM TG 263 initiative has published their recommendations on OAR and target nomenclature. The ETL user interface deploys this standardized nomenclature and requires the importer of the data to match the deemed OARs with their corresponding standard OAR and target names. In addition, this program also suggests a matching name based on an automated process of relabeling using our published techniques (OAR labels [19], radiomics features [20], and geometric information [21]). We find that these automated approaches provide an acceptable accuracy over the standard prostate and lung structure types. In order to gather the dose volume histogram data from the DICOM-RT dose and structure set files, we have deployed a DICOM-RT dosimetry parser software. If the DICOM-RT dose file exported by the TPS contains dose-volume histogram (DVH) information, we utilize it. However, if the file lacks this information, we employ our dosimetry parser software to calculate the DVH values from the dose and structure set volume information. | ||
===Mapping data to standardized terminology, data dictionary, and use of Semantic Web technologies=== | |||
For data to be interoperable, sharable outside the single [[hospital]] environment, and reusable for the various requirements of an LHS, the use of a standardized terminology and data dictionary is a key requirement. Specifically, clinical data should be transformed following the FAIR data principles. [22] An ontology describes a domain of classes and is defined as a conceptual model of knowledge representation. The use of ontologies and Semantic Web technologies plays a key role in transforming the healthcare data to be compatible with the FAIR principles. The use of ontologies enables the sharing of information between disparate systems within the multiple clinical domains. An ontology acts as a layer above the standardized data dictionary and terminology where explicit relationships—that is, predicates—are established between unique entities. Ontologies provide formal definitions of the clinical concepts used in the data sources and render the implicit meaning of the relationships among the different vocabulary and terminologies of the data sources explicitly. For example, it can be determined if two classes and data items found in different clinical databases are equivalent or if one is a subset of another. Semantic level information extraction and query are possible only with the use of ontology-based concepts of data mapping. | |||
A rapid way to look for new information on the internet is to use a search engine such as Google. These search engines return a list of suggested web pages devoid of context and semantics and require human interpretation to find useful information. The Semantic Web is a core technology that is used in order to organize and search for specific contextual information on the web. The Semantic Web, which is also known as Web 3.0, is an extension of the current World Wide Web (WWW) via a set of W3C data standards [23], with a goal to make internet data machine-readable instead of human-readable. For automatic processing of information by computers, Semantic Web extensions enable data (e.g., text, meta data on images, videos, etc.) to be represented with well-defined data structures and terminologies. To enable the encoding of semantics with the data, web technologies such as [[Resource Description Framework]] (RDF), Web Ontology Language (OWL), SPARQL Protocol, and RDF Query Language are used. RDF is a standard for sharing data on the web. | |||
We utilized an existing ontology known as Radiation Oncology Ontology (ROO) [[24]], available on the NCBO Bioportal website. [25] The main role of ROO is to define a broad coverage of main concepts used in the radiation oncology domain. The ROO currently consists of 1,183 classes with 211 predicates that are used to establish relationships between these classes. Upon inspection of this ontology, we noticed that the collection of classes and properties were missing some critical clinical elements such as smoking history, CTCAE v5 toxicity scores, diagnostic procedures such as Gleason scores, prostate-specific antigen (PSA) levels, patient reported outcome measures, Karnofsky performance status (KPS) scales, and radiation treatment modality. We utilized the ontology editor tool Protégé [26] for adding these key classes and properties in the updated ontology file. We reused entries from other published ontologies such as the [[National Cancer Institute]]'s ''NCI Thesaurus'' [27], [[International Statistical Classification of Diseases and Related Health Problems|International Classification of Diseases]] version 10 (ICD-10) [28], and Dbpedia [29] ontologies. We added 216 classes (categories defined in Table 1) with 19 predicate elements to the ROO. With over 100,000 terms, the ''NCI Thesaurus'' includes wide coverage of cancer terms, as well as mapping with external terminologies. The ''NCI Thesaurus'' is a product of NCI Enterprise Vocabulary Services (EVS), and its vocabularies consists of public information on cancer, definitions, synonyms, and other information on almost 10,000 cancers and related diseases, 17,000 single agents and related substances, as well as other topics that are associated with cancer. The list of high-level data categories, elements, and codes that are utilized in our work are included in the appendix (Appendix A2). | |||
{| | |||
| style="vertical-align:top;" | | |||
{| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="100%" | |||
|- | |||
| colspan="2" style="background-color:white; padding-left:10px; padding-right:10px;" |'''Table 1.''' Additional classes added to the Radiation Oncology Ontology (ROO) and used for mapping with our dataset. | |||
|- | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |Categories | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;" |# of classes | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Race, ethnicity | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |5 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Tobacco use | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |4 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Blood pressure + vitals | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |3 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Laboratory tests (e.g., creatinine, GFR, etc.) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |20 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Prostate-specific diagnostic tests (e.g., Gleason score, PSA, etc.) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |10 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Patient-reported outcome | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |8 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |CTCAE v5 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |152 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Therapeutic procedures (e.g., immunotherapy, targeted therapy, etc.) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |6 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Radiation treatment modality (e.g., photon, electron, proton, etc.) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |7 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Units (cGy) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |1 | |||
|- | |||
|} | |||
|} | |||
To use and validate the defined ontology, we mapped our data housed in the clinical data warehouse relational database with the concepts and relationships listed in the ontology. This mapping process linked each component (e.g., column headers, values) of the SQL relational database to its corresponding clinical concept (e.g., classes, relationships, and properties) in the ontology. To perform the mapping, the SQL database tables are analyzed and matched with the relevant concepts and properties in the ontology. This can be achieved by identifying the appropriate classes and relationships that best represent the data elements from the SQL relational database. For example, if the SQL relational table provides information about a patient's smoking history, the mapping process would identify the corresponding class or property in the ontology that represents smoking history. | |||
A correspondence between the table columns in the relational database and ontology entities was established using the D2RQ mapping script. An example of this mapping script is shown in Figure 2. With the use of the D2RQ mapping script, individual table columns in relational database schema were mapped to RDF ontology-based codes. This mapping script is executed by the D2RQ platform that connects to the SQL database, reads the schema, performs the mapping, and generates the output file in turtle syntax. Each SQL table column name is mapped to its corresponding class using the <tt>d2rq:ClassMap</tt> command. These classes are also mapped to existing ontology-based concept codes such as NCIT:C48720 for T1 staging. In order to define the relationships between two classes, the <tt>d2rq:refersToClassMap</tt> command is used. The properties of the different classes are defined using the <tt>d2rq:PropertyBridge</tt> command. | |||
Unique resource identifiers (URIs) are used for each entity for enabling the data to be machine-readable and for linking with other RDF databases. The mapping process is specific to the structure and content of the ontology being used, in this case, ROO. It relies on the defined classes, properties, and relationships within the ontology to establish the mapping between the SQL tables input data and the ontology terminology. While the mapping process is specific to the published ontology, it can potentially be generalized to other clinics or healthcare settings that utilize similar ontologies. The generalizability depends on the extent of similarity and overlap between the ontology being used and the terminologies and concepts employed in other clinics. If the ontologies share similar structures and cover similar clinical domains, the mapping process can be applied with appropriate adjustments to accommodate the specific terminologies and concepts used in the target clinic. | |||
[[File:Fig2 Kapoor JofAppCliMedPhys2023 24-10.jpg|900px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="900px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 2.''' Overview of the data mapping between the relational RO-CDW database and the hierarchical graph-based structure based on the defined ontology. The top rectangle displays an example of the various classes of the ontology and their relationships, including the NCI Thesaurus and ICD-10 codes. The bottom rectangle shows the relational database table, and the solid arrows between the top and bottom rectangles display the data mapping.</blockquote> | |||
|- | |||
|} | |||
|} | |||
===Importing data in knowledge based graph-based database=== | |||
The output file from the D2RQ mapping step is in Terse RDF Triple Language (turtle) syntax. This syntax is used for representing data in the semantic triples, which comprise a subject, predicate, and object. Each item in the triple is expressed as a Web URI. In order to search data from such formatted datasets, the dataset is imported in RDF knowledge graph databases. An RDF database, also called a Triplestore, is a type of graph database that stores RDF triples. The knowledge on the subject is represented in these triple formats consisting of subject, predicate, and object. An RDF knowledge graph can also be defined as labeled multi-diagraphs, which consist of a set of nodes which could be URIs or literals containing raw data, and the edges between these nodes represent the predicates. [30] The language used to reach data is called SPARQL—Query Language for RDF. It contains ontologies that are schema models of the database. Although SPARQL adopts various structures of SQL query language, SPARQL uses navigational-based approaches on the RDG graphs to query the data, which is quite different than the table-join-based storage and retrieval methods adopted in relational databases. In our work, we utilized the Ontotext GraphDB software [31] as our RDF store and SPARQL endpoint. | |||
===Ontology keyword-based searching tool=== | |||
It is common practice amongst healthcare providers to use different medical terms to refer to the same clinical concept. For example, if the user is searching for patient records that had a “heart attack,” then besides this text word search, they should also search for synonym concepts such as “myocardial infarction,” “acute coronary syndrome,” and so on. Ontologies such as ''NCI Thesaurus'' have listed synonym terms for each clinical concept. To provide an effective method to search the graph database, we built an ontology-based keyword search engine that utilizes the synonym-based term-matching methods. Another advantage of using ontology-based term searching is realized by using the class parent-children relationships. Ontologies are hierarchical in nature, with the terms in the hierarchy often forming a directed acyclic graph (DAG). For example, if we are searching for patients in our database with clinical stage T1, the matching patient list will only comprise patients that have T1 stage ''NCI Thesaurus'' code (NCIT: C48720) in the graph database. These matching patients will not return any patients with T1a, T1b, and T1c sub-categories that are children of the parent T1 staging class. We built this search engine where we can search on any clinical term and its matching patient records based on both parent and children classes, which are abstracted. | |||
The method that is used in this search engine is as follows. When the user wants to use the ontology to query the graph-based medical records, the only input necessary is the clinical query terms (q-terms) and an indication of whether the synonyms should also be considered while retrieving the patient records. The user has the option to specify the multiple levels of child class search and parent classes to be included in the search parameters. The software will then connect to the Bioportal database via REST API and perform the search to gather the matching classes for the q-terms and the options specified in the program. Using the list of matching classes, a SPARQL-based query is generated and executed with our patient graph database and matching patient list, and the q-term based clinical attributes are returned to the user. | |||
In order to find patients that have not the same but similar attributes based on the search parameters, we have designed a patient similarity search method. The method employed to identify similar patients based on matching knowledge graph attributes involves the creation of a text corpus by performing breadth-first search (BFS) random walks on each patient's individual knowledge graph. This process allows us to explore the graph structure and extract the necessary information for analysis. Within each patient's knowledge graph, approximately 18−25 categorical features were extracted in the text corpus. It is important to note that the number of features extracted from each patient may vary, as it depends on the available data and the complexity of the patient's profile. These features included the diagnosis; tumour, node, metastasis (TNM) staging; [[histology]]; smoking status; performance status; [[pathology details]]; radiation treatment modality; technique; and toxicity grades. | |||
This text corpus is then used to create word embeddings that can be used later to search for similar patients based on similarity and distance metrics. We utilized four vector embedding models, namely Word2Vec [32], Doc2Vec [33], GloVe [34], and FastText [35] to train and generate vector embeddings. The output of word embedding models are vectors, one for each word in the training dictionary, that effectively capture relationships between words. The architecture of these word embedding models is based on a single hidden layer neural network. The description of these models is provided in Appendix A1. | |||
The text corpus used for training is obtained from the Bioportal website, which encompasses NCIT, ICD, and SNOMED codes, as well as class definition text, synonyms, hyponyms terms, parent classes, and sibling classes. We scraped 139,916 classes from the Bioportal website using API calls and used this dataset to train our word embedding models. By incorporating this diverse and comprehensive dataset, we aimed to capture the semantic relationships and contextual information relevant to the medical domain. The training process involved iterating over the training dataset for a total of 100 epochs using CPU hardware. During training, the models learned the underlying patterns and semantic associations within the text corpus, enabling them to generate meaningful vector representations for individual words, phrases, or documents. Once the models were trained, we utilized them to generate vector embeddings for the individual patient text corpus that we had previously obtained. These embeddings served as numerical representations of the patient data, capturing the semantic and contextual information contained within the patient-specific text corpus. The Cosine similarity, Euclidean distance, Manhattan distance, and Minkowski distance metrics are employed to measure the distance between the matched patients and all patient feature vectors. | |||
Figure 3 shows the design architecture of the software system. The main purpose of this search engine is to provide the users with a simple interface to search the patient records. | |||
[[File:Fig3 Kapoor JofAppCliMedPhys2023 24-10.jpg|1100px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="1100px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 3.''' Design architecture for the ontology-based keyword search system. When the user wants to query the patient graph database to retrieve matching records, the only input necessary is the medical terms (q-terms) and an indication to include any synonym, parent, or children terminology classes in the search. The software queries the Bioportal API and retrieves all the matching NCIT, SNOMED, ICD-10 classes to the q-terms. A SPARQL query is generated and executed on the graph database SPARQL endpoint, and the results indicating the matching patient records and their corresponding data fields are displayed to the user. Our architecture includes the generation of text corpus from breadth-first search (BFS) of individual patient graphs and using word embedding models to generate feature vectors to identify similar patient cohorts.</blockquote> | |||
|- | |||
|} | |||
|} | |||
==Results== | |||
===Mapping data to the ontology=== | |||
With the aim of testing out the data pipeline and infrastructure, we used our clinical database that has 1,660 patient clinical and dosimetry records. These records are from patients treated with radiotherapy for prostate cancer, non-small cell lung cancer, and small cell lung cancer disease. There are 35,303 clinical and 12,565 DVH based data elements that are stored in our RO-CDW database for these patients. All these data elements were mapped to the ontology using the D2RQ mapping language, resulting in 504,180 RDF tuples. In addition to the raw data, these tuples also defined the interrelationships amongst various defined classes in the dataset. An example of the output RDF tuple file is shown in Figure 4, displaying the patient record relationship with diagnosis, TNM staging, etc. All the entities and predicates in the output RDF file have a URI, which is resolvable as a link for the computer program or human to gather more data on the entities or class. For example, the RDF viewer would be able to resolve the address <nowiki>http://purl.obolibrary.org/obo/NCIT_48720</nowiki> to gather details on the T-stage such as concept definitions, synonym, relationship with other concepts and classes, etc. | |||
We were able to achieve a mapping completeness of 94.19% between the records in our clinical database and RDF tuples. During the validation process, we identified several ambiguities or inconsistencies in the data housed in the relational database, such as indication of use of ECOG instrument for performance status evaluation but missing values for ECOG performance status score, record of T stage but nodal and metastatic stage missing, and delivered number fractions missing with the prescribed dose information. To maintain data integrity and accuracy, the D2RQ mapping script was designed to drop these values due to missing or incomplete data or ambiguous information. Additionally, the validation process thoroughly examined the interrelationships among the defined classes in the dataset. We verified that the relationships and associations between entities in the RDF tuples accurately reflected the relationships present in the original clinical data. Any discrepancies or inconsistencies found during this analysis were identified and addressed to ensure the fidelity of the mapped data. To evaluate the accuracy of the mapping process, we conducted manual spot checks on a subset of the RDF tuples. This involved randomly selecting samples of RDF tuples and comparing the mapped values to the original data sources. Through these spot checks, we ensured that the mapping process accurately represented and preserved the information from the clinical and dosimetry data during the transformation into RDF tuples. Overall, the validation process provided assurance that the pipeline effectively transformed the clinical and dosimetry data stored in the RO-CDW database into RDF tuples while preserving the integrity, accuracy, and relationships of the original data. | |||
[[File:Fig4 Kapoor JofAppCliMedPhys2023 24-10.jpg|900px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="900px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 4.''' Example of the output RDF tuple file.</blockquote> | |||
|- | |||
|} | |||
|} | |||
===Visualization of data in ontology-based graphical format=== | |||
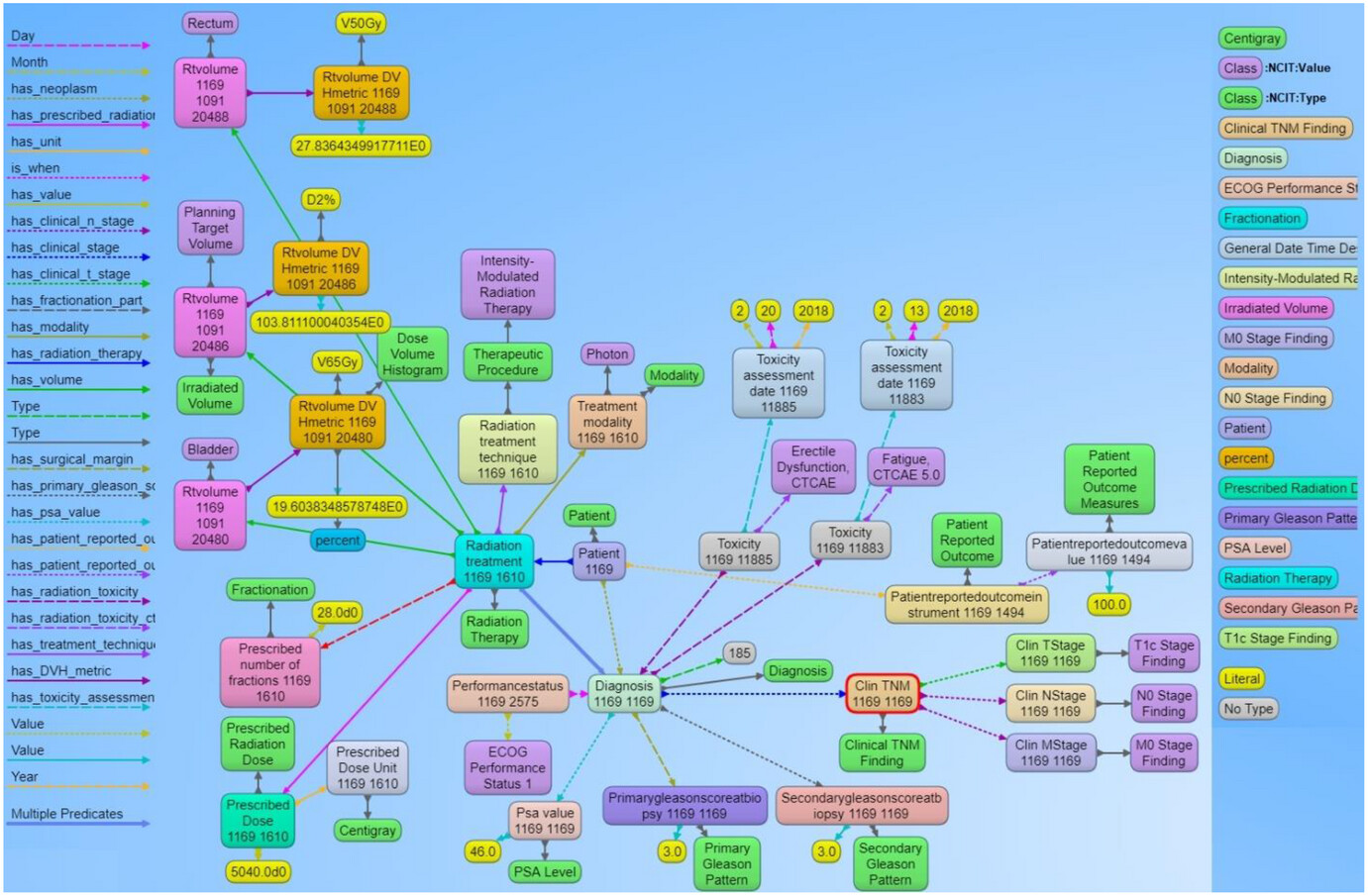
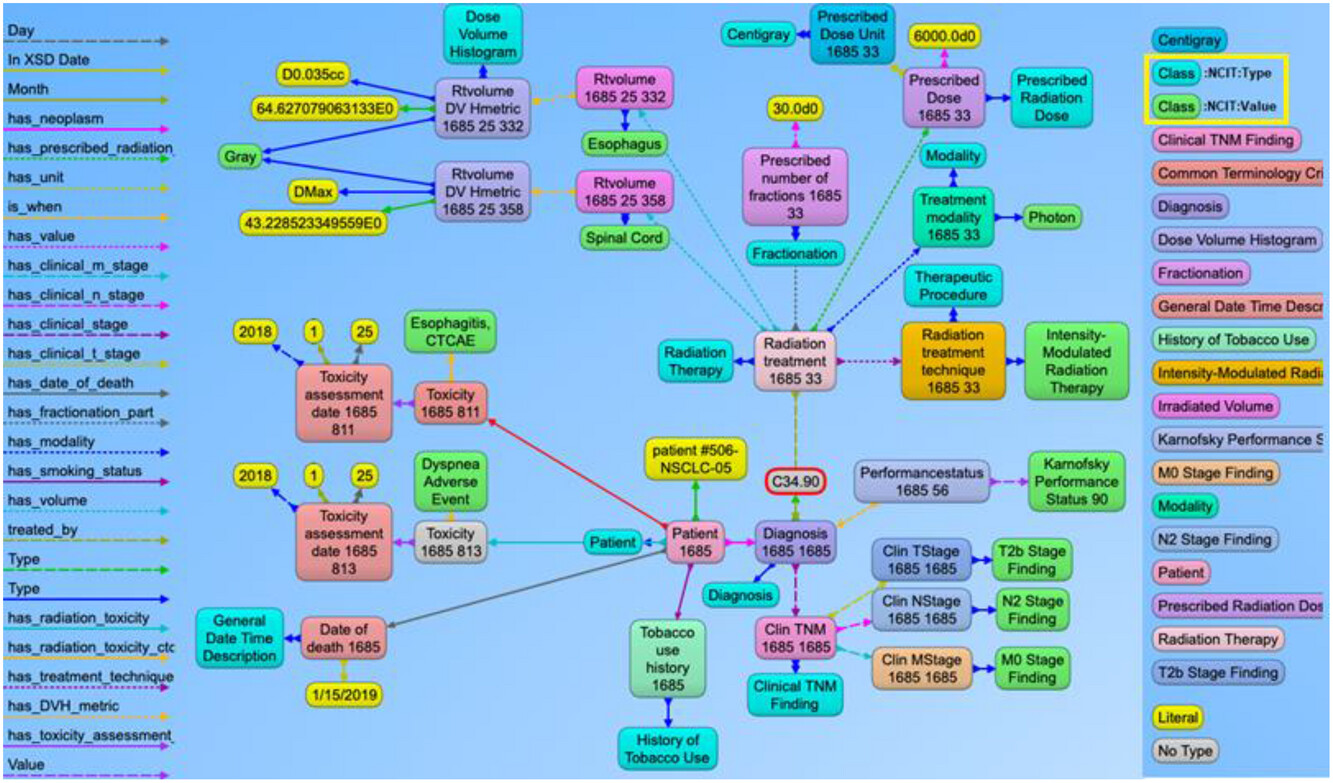
Visualizations on ontologies play a key role for users to understand the structure of the data and work with the dataset and its applications. This has an appealing potential when it comes to exploring or verifying complex and large collections of data such as ontologies. We utilized the Allegrograph Gruff toolkit [36] that enables users to create visual knowledge graphs that display data relationships in a neat graphical user interface (GUI). The Gruff toolkit uses simple SPARQL queries to gather the data for rendering the graph with nodes and edges. These visualizations are useful because they increase the users’ understanding of data by instantly illustrating relevant relationships amongst class and concepts, hidden patterns, and data significance to outcomes. An example of the graph-based visualization for a prostate and non-small cell lung cancer patient is shown in Figures 5 and 6. Here all the nodes stand for concepts and classes, and the edges represent relationships between these concepts. All the nodes in the graph have URIs that are resolvable as a web link for the computer program or human to gather more data on the entities or classes. The color of the nodes in the graph visualization are based on the node type, and there are inherent properties of each node that include the unique system code (e.g., NCIT code or ICD code, etc.), synonyms terms, definitions, value type (e.g., string, integer, floating point number, etc.). The edges connecting the nodes are defined as properties and stored as predicates in the ontology data file. The use of these predicates enables the computer program to effectively find the queried nodes and their interrelationships. Each of these properties are defined with URIs that are available for gathering more detailed information on the relationship definitions. The left panel in Figures 5 and 6 shows various property types or relationship types that connect the nodes in the graph. Using SPARQL language and Gruff visualization tools, users can query the data without having any prior knowledge of the relational database structure or schema, since these SPARQL queries are based on universal publish classes defined in the ''NCI Thesaurus'', Units Ontology, and ICD-10 ontologies. | |||
[[File:Fig5 Kapoor JofAppCliMedPhys2023 24-10.jpg|1100px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="1100px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 5.''' Example of the graph structure of a prostate cancer patient record based on the ontology. Each node in the graph are entities that represent objects or concepts and have a unique identifier and can have properties and relationships to other nodes in the graph. These nodes are connected by directed edges, representing relationships between the information, such as the relationship between the diagnosis node and the radiation treatment node. Similarly, there are edges from the diagnosis node to the toxicity node and further to the specific CTCAE toxicity class, indicating that the patient was evaluated for adverse effects after receiving radiation therapy. The different types of edge relationships from the ontology that are used in this example are listed on the left panel of the figure. The right panel shows different types of nodes that are used in the example.</blockquote> | |||
|- | |||
|} | |||
|} | |||
[[File:Fig6 Kapoor JofAppCliMedPhys2023 24-10.jpg|1100px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="1100px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 6.''' Example of the graph structure of a non-small cell lung cancer (NSCLC) patient based on the ontology. This has a similar structure to the previous prostate cancer example with NSCLC content. The nodes in green and aqua blue color (highlighted in the right panel) indicate the use of NCI Thesaurus classes to represent the use of standard terminology to define the context for each node present in the graph. For simpler visualization, the NCI Thesaurus codes and URIs are not displayed with this example.</blockquote> | |||
|- | |||
|} | |||
|} | |||
Finally, these SPARQL queries can be used with commonly available programming languages like Python and [[R (programming language)|R]] via REST APIs. We also verified data from the SPARQL queries and the SQL queries from the CDW database for accuracy of the mapping. Our analysis found no difference in the resultant data from the two query techniques. The main advantage of using the SPARQL method is that the data can be queried without any prior knowledge of the original data structure based on the universal concepts defined in the ontology. Also, the data from multiple sources can be seamlessly integrated in the RDF graph database without the use of complex data matching techniques and schema modifications, which is currently required with relational databases. This is only possible if all the data stored in the RDF graph database refers to published codes from the commonly used ontologies. | |||
===Searching the data using ontology-based keywords=== | |||
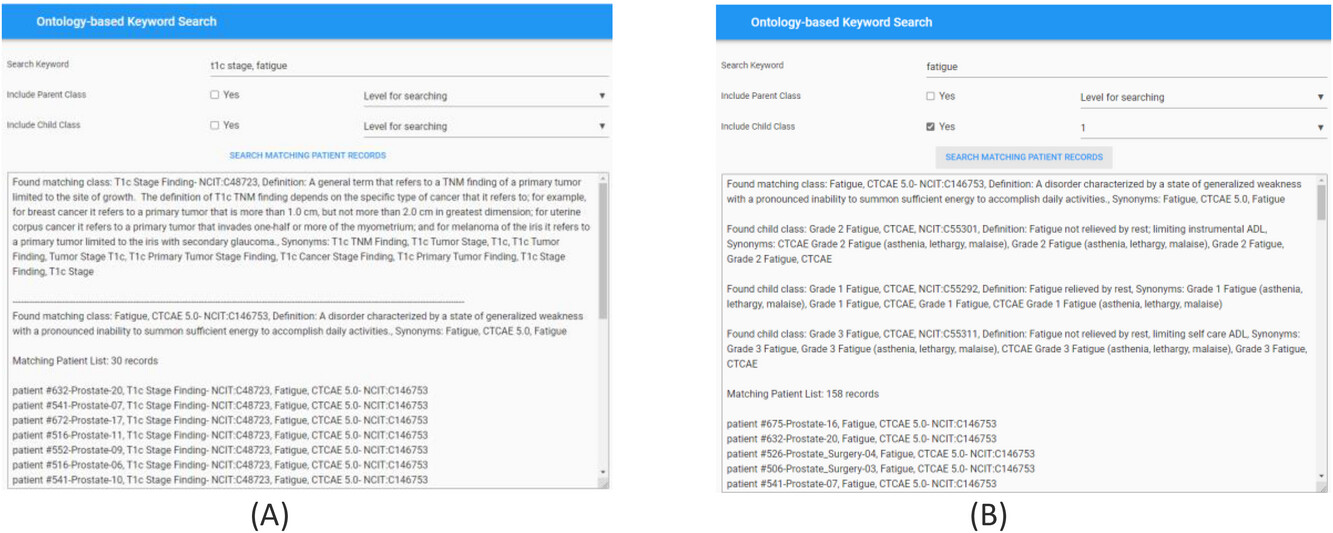
For effective searching of discrete data from the RDF graph database, we built an ontology-based keyword searching web tool. The public website for this tool is [https://hinge-ontology-search.anvil.app https://hinge-ontology-search.anvil.app]. Here we are able to search the database based on keywords (q-terms). The tool is connected to the Bioportal via REST API and finds the matching classes or concepts and renders the results including the class name, NCIT code, and definitions. We specifically used the ''NCI Thesaurus'' ontology for our query which is 112 MB in size and contains approximately 64,000 terms. The search tool can find the classes based on synonym term queries where it matches the q-terms with the listed synonym terms in the classes (Figure 7a). The tool has features to search the child and parent classes on the matching q-term classes. A screenshot of the web tool with the child class search is shown in Figure 7b. The user can also specify the level of search ,which indicates if the returned classes should include classes of children of children. In the example in Figure 9b, we are showing the q-term used for searching “fatigue” while including the child classes up to one level, and the return classes included the fatigue based CTCAE class and the grade 1, 2, 3 fatigue classes. Once all the classes used for searching are found by the tool, it searches the RDF graph database for matching patient cases with these classes. The matching patient list, including the found class in the patient's graph, is displayed to the user. This tool is convenient for end users to abstract cohorts of patients that have particular classes or concepts in their records without the user learning and implementing the complex SPARQL query language. Based on our evaluation, we found that the average time taken to obtain results is less than five seconds per q-term if there are less than five child classes in the query. The maximum time taken is 11 seconds for a q-term that had 16 child classes. | |||
[[File:Fig7 Kapoor JofAppCliMedPhys2023 24-10.jpg|1200px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="1200px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 7.''' Screenshot of the ontology-based keyword search portal. '''(A)''' Search performed using two q-terms returns results with definitions of the matching classes from the Bioportal and the corresponding patient records from the RDF graph database. '''(B)''' Search performed to include child class up to one level on the matching q-term class. Returned results display the matching class, child classes with Fatigue CTCAE grades, and matching patient records from the RDF graph database.</blockquote> | |||
|- | |||
|} | |||
|} | |||
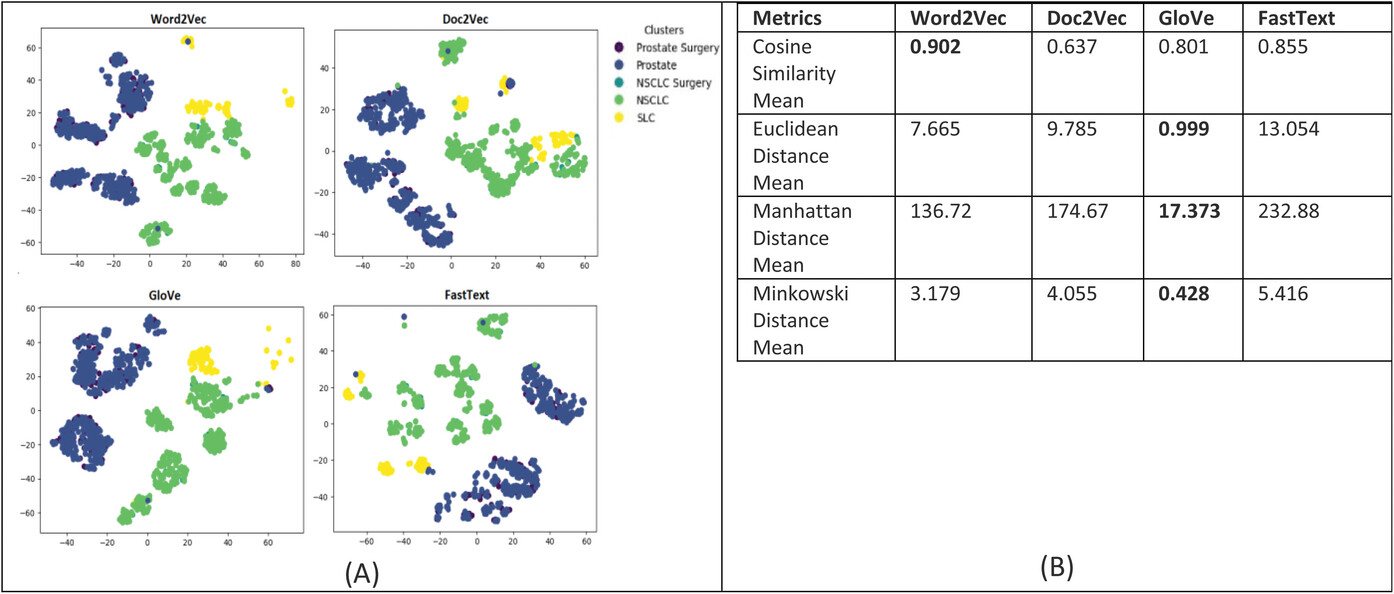
For evaluating the patient similarity-based work embedding models, we evaluated the quality of the feature embedding-based vectors produced by using the technique called t-Distributed Stochastic Neighbor Embedding (t-SNE) and cluster analysis with a predetermined number of clusters set to five based on the diagnosis groups for our patient cohort. Our main objective is to determine the similarity between patient data that are in the same cluster based on their corresponding diagnosis groups. This method can reveal the local and global features encoded by the feature vectors and thus can be used to visualize clusters within the data. We applied t-SNE to all 1,660 patient feature-based vectors produced via the four word embedding models. The t-SNE plot is shown in Figure 8; it shows that the disease data points can be grouped into five clusters with varying degrees of separability and overlap. The analysis of patient similarity using different embedding models revealed interesting patterns. The Word2Vec model showed the highest mean cosine similarity of 0.902, indicating a relatively higher level of similarity among patient embeddings within the five diagnosis groups. In contrast, the Doc2Vec model exhibited a lower mean cosine similarity of 0.637. The GloVe model demonstrated a moderate mean cosine similarity of 0.801, while the FastText model achieved a similar level of 0.855. Regarding distance metrics, the GloVe model displayed lower mean Euclidean and Manhattan distances, suggesting that patient embeddings derived from this model were more compact and closer in proximity. Conversely, the Doc2Vec, Word2Vec, and FastText models yielded higher mean distances, indicating greater variation and dispersion among the patient embeddings. These findings provide valuable insights into the performance of different embedding models for capturing patient similarity, facilitating improved understanding and decision-making in the clinical domain. | |||
[[File:Fig8 Kapoor JofAppCliMedPhys2023 24-10.jpg|1200px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="1200px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 8.''' '''(A)''' Annotation embeddings produced by Word2Vec, Doc2Vec, GloVe, and FastText, a 2D-image of the embeddings projected down to three dimensions using the T-SNE technique. Each point indicates one patient, and color of a point indicates the cohort of the patient based on the diagnosis-based cluster. A good visualization result is that the points of the same color are near each other. '''(B)''' Results of the evaluation metrics used to measure patient similarity. The Word2Vec model had the best cosine similarity, and the GloVe model had the best Euclidean, Manhattan, and Minkowski distance, suggesting that patient embeddings derived from this model were more compact and closer in proximity.</blockquote> | |||
|- | |||
|} | |||
|} | |||
==Discussion== | |||
Revision as of 00:43, 10 May 2024
| Full article title | Infrastructure tools to support an effective radiation oncology learning health system |
|---|---|
| Journal | Journal of Applied Clinical Medical Physics |
| Author(s) | Kapoor, Rishabh; Sleeman IV, William C.; Ghosh, Preetam; Palta, Jatinder |
| Author affiliation(s) | Virginia Commonwealth University |
| Primary contact | rishabh dot kapoor at vcuhealth dot org |
| Year published | 2023 |
| Volume and issue | 24(10) |
| Article # | e14127 |
| DOI | 10.1002/acm2.14127 |
| ISSN | 1526-9914 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://aapm.onlinelibrary.wiley.com/doi/10.1002/acm2.14127 |
| Download | https://aapm.onlinelibrary.wiley.com/doi/pdfdirect/10.1002/acm2.14127 (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Purpose: The concept of the radiation oncology learning health system (RO‐LHS) represents a promising approach to improving the quality of care by integrating clinical, dosimetry, treatment delivery, and research data in real‐time. This paper describes a novel set of tools to support the development of an RO‐LHS and the current challenges they can address.
Methods: We present a knowledge graph‐based approach to map radiotherapy data from clinical databases to an ontology‐based data repository using FAIR principles. This strategy ensures that the data are easily discoverable, accessible, and can be used by other clinical decision support systems. It allows for visualization, presentation, and analysis of valuable data and information to identify trends and patterns in patient outcomes. We designed a search engine that utilizes ontology‐based keyword searching and synonym‐based term matching that leverages the hierarchical nature of ontologies to retrieve patient records based on parent and children classes, as well as connects to the Bioportal database for relevant clinical attributes retrieval. To identify similar patients, a method involving text corpus creation and vector embedding models (Word2Vec, Doc2Vec, GloVe, and FastText) are employed, using cosine similarity and distance metrics.
Results: The data pipeline and tool were tested with 1,660 patient clinical and dosimetry records, resulting in 504,180 RDF (Resource Description Framework) tuples and visualized data relationships using graph‐based representations. Patient similarity analysis using embedding models showed that the Word2Vec model had the highest mean cosine similarity, while the GloVe model exhibited more compact embeddings with lower Euclidean and Manhattan distances.
Conclusions: The framework and tools described support the development of an RO‐LHS. By integrating diverse data sources and facilitating data discovery and analysis, they contribute to continuous learning and improvement in patient care. The tools enhance the quality of care by enabling the identification of cohorts, clinical decision support, and the development of clinical studies and machine learning (ML) programs in radiation oncology.
Keywords: FAIR, learning health system infrastructure, ontology, Radiation Oncology Ontology, Semantic Web
Background and significance
For the past three decades, there is a growing interest in building learning organizations to address the most pressing and complex business, social, and economic challenges facing society today. [1] For healthcare, the National Academy of Medicine has defined the concept of a learning health system (LHS) as an entity where science, incentive, culture, and informatics are aligned for continuous innovation, with new knowledge capture and discovery as an integral part for practicing evidence-based medicine. [2] The current dependency on randomized controlled clinical trials that use a controlled environment for scientific evidence creation with only a small percent (<3%) of patient samples is inadequate now and may be irrelevant in the future since these trials take too much time, are too expensive, and are fraught with questions of generalizability. The Agency for Healthcare Research and Quality has also been promoting the development of LHSs as part of a key strategy for healthcare organizations to make transformational changes to improve healthcare quality and value. Large-scale healthcare systems are now recognizing the need to build infrastructure capable of continuous learning and improvement in delivering care to patients and address critical population health issues. [3] In an LHS, data collection should be performed from various sources such as electronic health records (EHRs), treatment delivery records, imaging records, patient-generated data records, and administrative and claims data, which then allows for this aggregated data to be analyzed for generating new insights and knowledge that can be used to improve patient care and outcomes.
However, only a few attempts at leveraging existing infrastructure tools used in routine clinical practice to transform the healthcare domain into an LHS have been made. [5, 6] Some examples of actual implementation have emerged, but by and large these concepts have been mostly discussed as conceptual ideas and strategies in the literature. There are several data organization and management challenges that must be addressed in order to effectively implement a radiation oncology LHS:
- 1. Data integration: Radiation oncology data are generated from a variety of sources, including EHRs, imaging systems, treatment planning systems (TPSs), and clinical trials. Integration of this data into a single repository can be challenging due to differences in data formats, terminologies, and storage systems. There is often significant semantic heterogeneity in the way that different clinicians and researchers use terminology to describe radiation oncology data. For example, different institutions may use different codes or terms to describe the same condition or treatment.
- 2. Data stored in disparate database schemas: Presently, the EHR, TPS, and treatment management system (TMS) data are housed in a series of relational database management systems (RDMS), which have rigid database structures, varying data schemas and can include lots of uncoded textual data. Tumor registries also stores data in their own defined schemas. Although the column names in the relational databases between two software products might be the same, semantic meaning based on the application of use may be completely different. Changing a database schema requires a lot of programming effort and code changes because of the rigid structure of the stored data, and it is generally advisable to retire old tables and build new tables with the added column definitions.
- 3. Episodic linking of records: Episodic linking of records refers to the process of integrating patient data from multiple encounters or episodes of care into a single comprehensive record. This record includes information about the patient's medical history, diagnosis, treatment plan, and outcomes, which can be used to improve care delivery, research, and education. Linking multiple data sources based on the patients episodic history of care is quite challenging because the heterogeneity of these data sources does not normally follow any common data storage standards.
- 4. Build data query tools based on semantic meaning of the data: Since the data are currently stored in multiple RDMSs for the specific purpose to cater the operations aspects of the patient care, extracting common semantic meaning from this data is very challenging. Common semantic meaning in healthcare data is typically achieved through the use of standardized vocabularies and ontologies that define concepts and relationships between them. Developing data query tools based on semantic meaning requires a high level of expertise in both the technical and domain-specific aspects of radiation oncology. Moreover, executing complex data queries, which includes tree-based queries, recursive queries, and derived data queries requires multiple tables joining operations in RDMSs, which is a costly operation.
While we are on the cusp of an artificial intelligence (AI) revolution in biomedicine, with the fast-growing development of advanced machine learning (ML) methods that can analyze complex datasets, there is an urgent need for a scalable intelligent infrastructure that can support these methods. The radiation oncology domain is also one of the most technically advanced medical specialties, with a long history of electronic data generation (e.g., radiation treatment (RT) simulation, treatment planning, etc.) that is modeled for each individual patient. This large volume of patient-specific real-world data captured during routine clinical practice, dosimetry, and treatment delivery make this domain ideally suited for rapid learning. [4] Rapid learning concepts could be applied using an LHS, providing the potential to improve patient outcomes and care delivery, reduce costs, and generate new knowledge from real world clinical and dosimetry data.
Several research groups in radiation oncology, including the University of Michigan, MD Anderson, and Johns Hopkins, have developed data gathering platforms with specific goals. [5] These platforms—such as the M-ROAR platform [6] at the University of Michigan, the system-wide electronic data capture platform at MD Anderson [7], and the Oncospace program at Johns Hopkins [8]—have been deployed to collect and assess practice patterns, perform outcome analysis, and capture RT-specific data, including dose distributions, organ-at-risk (OAR) information, images, and outcome data. While these platforms serve specific purposes, they rely on relational database-based systems without utilizing standard ontology-based data definitions. However, knowledge graph-based systems offer significant advantages over these relational database-based systems. Knowledge graph-based systems provide a more integrated and comprehensive representation of data by capturing complex relationships, hierarchies, and semantic connections between entities. They leverage ontologies, which define standardized and structured knowledge, enabling a holistic view of the data and supporting advanced querying and analysis capabilities. Furthermore, knowledge graph-based systems promote data interoperability and integration by adopting standard ontologies, facilitating collaboration and data sharing across different research groups and institutions. As such, knowledge graph-based systems are able to help ensure that research data is more findable, accessible, interoperable, and reusable (FAIR). [22]
In this paper, we set out to contribute to the advancement of the science of LHSs by presenting a detailed description of the technical characteristics and infrastructure that were employed to design a radiation oncology LHS specifically with a knowledge graph approach. The paper also describes how we have addressed the challenges that arise when building such a system, particularly in the context of constructing a knowledge graph. The main contributions of our work are as follows:
- 1. Provides an overview of the sources of data within radiation oncology (EHRs, TPS, TMS) and the mechanism to gather data from these sources in a common database.
2. Maps the gathered data to a standardized terminology and data dictionary for consistency and interoperability. Here we describe the processing layer built for data cleaning, checking for consistency and formatting before the extract, transform, and load (ETL) procedure is performed in a common database.
3. Adds concepts, classes, and relationships from existing NCI Thesaurus and SNOMED terminologies to the previously published Radiation Oncology Ontology (ROO) to fill in gaps with missing critical elements in the LHS.
4. Presents a knowledge graph visualization that demonstrates the usefulness of the data, with nodes and relationships for easy understanding by clinical researchers.
5. Develops an ontology-based keyword searching tool that utilizes semantic meaning and relationships to search the RDF knowledge graph for similar patients.
6. Provides a valuable contribution to the field of radiation oncology by describing an LHS infrastructure that facilitates data integration, standardization, and utilization to improve patient care and outcomes.
Material and methods
Gather data from multiple source systems in the radiation oncology domain
The adoption of EHRs in patients' clinical management is rapidly increasing in healthcare, but the use of data from EHRs in clinical research is lagging. The utilization of patient-specific clinical data available in EHRs has the potential to accelerate learning and bring value in several key topics of research, including comparative effectiveness research, cohort identification for clinical trial matching, and quality measure analysis. [9, 10] However, there is an inherent lack of interest in the use of data from the EHR for research purposes since the EHR and its data were never designed for research. Modern EHR technology has been optimized for capturing health details for clinical record keeping, scheduling, ordering, and capturing data from external sources such as laboratories, diagnostic imaging, and capturing encounter information for billing purposes. [11] Many data elements collected in routine clinical care, which are critical for oncologic care, are neither collected as structured data elements nor with the same defined rigor as those in clinical trials. [12, 13]
Given all these challenges with using data from EHRs, we have designed and built a clinical software called Health Information Gateway Exchange (HINGE). HINGE is a web-based electronic structured data capture system that has electronic data sharing interfaces using the Fast Healthcare Interoperability Resources (FHIR) Health Level 7 (HL7) standards with a specific goal to collect accurate, comprehensive, and structured data from EHRs. [14] FHIR is an advanced interoperability standard introduced by standards developing organization HL7. FHIR is based on the previous HL7 standards (version 1 & 2) and provides a representational state transfer (REST) architecture, with an application programming interface (API) in Extensible Markup Language (XML) and JavaScript Object Notation (JSON) formats. Additionally, there has also been recent regulatory and legislative changes promoting the use of FHIR standards for interoperability and interconnectivity of healthcare systems. [16] HINGE has employed the FHIR interfaces with EHRs to retrieve required patient details such as demographics; list of allergies; prescribed active medications; vitals; lab results; surgery, radiology, and pathology reports; active diagnosis; referrals; encounters; and survival information. We have described the design and implementation of HINGE in our previous publication. [15] In summary, HINGE is designed to automatically capture and abstract clinical, treatment planning, and delivery data for cancer patients receiving radiotherapy. The system uses disease site-specific “smart” templates to facilitate the entry of relevant clinical information by physicians and clinical staff. The software processes the extracted data for quality and outcome assessment, using well-defined clinical and dosimetry quality measures defined by disease site experts in radiation oncology. The system connects seamlessly to the local IT/medical infrastructure via interfaces and cloud services and provides tools to assess variations in radiation oncology practices and outcomes, and determine gaps in radiotherapy quality delivered by each provider.
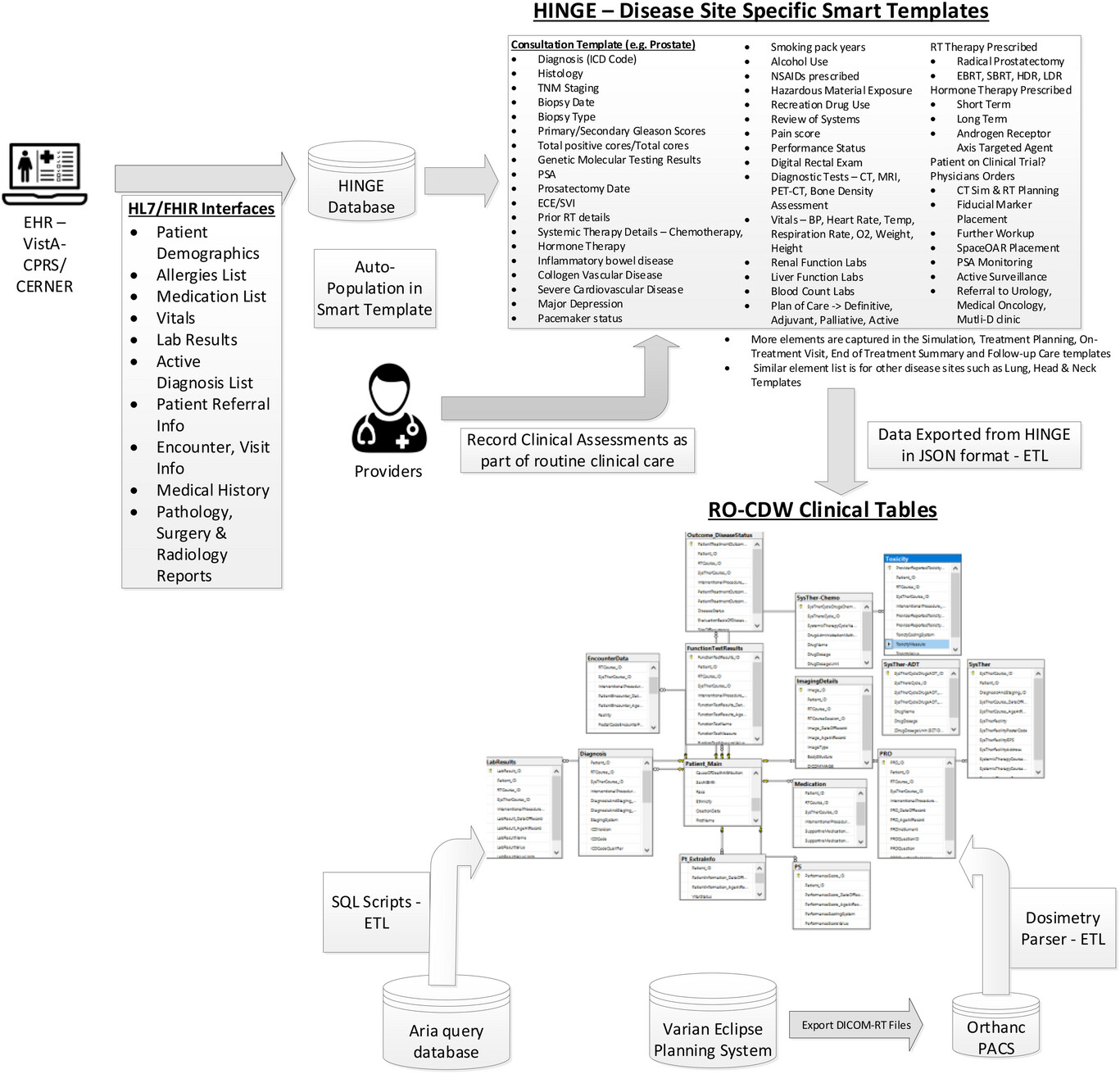
We created a data pipeline from HINGE to export discrete data in JSON-based format. These data are then fed to the extract, transform, and load (ETL) processor. An overview of the data pipeline is shown in Figure 1. ETL is a three-step process where the data are first extracted, transformed (i.e., cleaned, formatted), and loaded into an output radiation oncology clinical data warehouse (RO-CDW) repository. Since HINGE templates do not function as case report forms and they are formatted based on an operational data structure, the data cleaning process is performed with some basic data preprocessing, including cleaning and checking for redundancy in the dataset, ignoring null values while making sure each data element has its supporting data elements populated in the dataset. As there are several types of datasets, each dataset requires a different type of cleaning. Therefore, multiple scripts for data cleaning have been prepared. The following outlines some of the checks that have been performed using the cleaning scripts.
- 1. Data type validation: We verified whether the column values were in the correct data types (e.g., integer, string, float). For instance, the “Performance Status Value” column in a patient record should be an integer value.
- 2. Cross-field consistency check: Some fields require other column values to validate their content. For example, the “Radiotherapy Treatment Start Date” should not be earlier than the “Date of Diagnosis.” We conducted a cross-field validation check to ensure that such conditions were met.
- 3. Mandatory element check: Certain columns in the input data file cannot be empty, such as “Patient ID Number” and “RT Course ID” in the dataset. We performed a mandatory field check to ensure that these fields were properly filled.
- 4. Range validation: This check ensures that the values fall within an acceptable range. For example, the “Marital Status” column should contain values between 1 to 9.
- 5. Format check: We verified the format of data values to ensure that they were consistent with the expected year-month-day (YYYYMMDD) format.
|
The main purpose of this step is to ensure that the dataset is of high quality and fidelity when loaded in RO-CDW. In the data loading process, we have written SQL and .Net-based scripts to transform the data into RO-CDW-compatible schema and load them into a Microsoft SQL Server 2016 database. When the data are populated, unique identifiers are assigned to each data table entry, and interrelationships are maintained within the tables so that the investigators can use query tools to query and retrieve the data, identify patient cohorts, and analyze the data.
We have deployed a free, open-source, and light-weight DICOM server known as Orthanc17 to collect DICOM-RT datasets from any commercial TPS. Orthanc is a simple yet powerful standalone DICOM server designed to support research and provide query/retrieve functionality of DICOM datasets. Orthanc provides a RESTful API that makes it possible to program using any computer language where DICOM tags stored in the datasets can be downloaded in a JSON format. We used the Python plug-in to connect with the Orthanc database to extract the relevant tag data from the DICOM-RT files. Orthanc was able to seamlessly connect with the Varian Eclipse planning system with the DICOM DIMSE C-STORE protocol. [18] Since the TPS conforms to the specifications listed under the Integrating the Healthcare Enterprise—Radiation Oncology (IHE-RO) profile, the DICOM-RT datasets contained all the relevant tags that were required to extract data.
One of the major challenges with examining patients’ DICOM-RT data is the lack of standardized organs at risk (OAR) and target names, as well as ambiguity regarding dose-volume histogram metrics, and multiple prescriptions mentioned across several treatment techniques. With the goal of overcoming these challenges, the AAPM TG 263 initiative has published their recommendations on OAR and target nomenclature. The ETL user interface deploys this standardized nomenclature and requires the importer of the data to match the deemed OARs with their corresponding standard OAR and target names. In addition, this program also suggests a matching name based on an automated process of relabeling using our published techniques (OAR labels [19], radiomics features [20], and geometric information [21]). We find that these automated approaches provide an acceptable accuracy over the standard prostate and lung structure types. In order to gather the dose volume histogram data from the DICOM-RT dose and structure set files, we have deployed a DICOM-RT dosimetry parser software. If the DICOM-RT dose file exported by the TPS contains dose-volume histogram (DVH) information, we utilize it. However, if the file lacks this information, we employ our dosimetry parser software to calculate the DVH values from the dose and structure set volume information.
Mapping data to standardized terminology, data dictionary, and use of Semantic Web technologies
For data to be interoperable, sharable outside the single hospital environment, and reusable for the various requirements of an LHS, the use of a standardized terminology and data dictionary is a key requirement. Specifically, clinical data should be transformed following the FAIR data principles. [22] An ontology describes a domain of classes and is defined as a conceptual model of knowledge representation. The use of ontologies and Semantic Web technologies plays a key role in transforming the healthcare data to be compatible with the FAIR principles. The use of ontologies enables the sharing of information between disparate systems within the multiple clinical domains. An ontology acts as a layer above the standardized data dictionary and terminology where explicit relationships—that is, predicates—are established between unique entities. Ontologies provide formal definitions of the clinical concepts used in the data sources and render the implicit meaning of the relationships among the different vocabulary and terminologies of the data sources explicitly. For example, it can be determined if two classes and data items found in different clinical databases are equivalent or if one is a subset of another. Semantic level information extraction and query are possible only with the use of ontology-based concepts of data mapping.
A rapid way to look for new information on the internet is to use a search engine such as Google. These search engines return a list of suggested web pages devoid of context and semantics and require human interpretation to find useful information. The Semantic Web is a core technology that is used in order to organize and search for specific contextual information on the web. The Semantic Web, which is also known as Web 3.0, is an extension of the current World Wide Web (WWW) via a set of W3C data standards [23], with a goal to make internet data machine-readable instead of human-readable. For automatic processing of information by computers, Semantic Web extensions enable data (e.g., text, meta data on images, videos, etc.) to be represented with well-defined data structures and terminologies. To enable the encoding of semantics with the data, web technologies such as Resource Description Framework (RDF), Web Ontology Language (OWL), SPARQL Protocol, and RDF Query Language are used. RDF is a standard for sharing data on the web.
We utilized an existing ontology known as Radiation Oncology Ontology (ROO) 24, available on the NCBO Bioportal website. [25] The main role of ROO is to define a broad coverage of main concepts used in the radiation oncology domain. The ROO currently consists of 1,183 classes with 211 predicates that are used to establish relationships between these classes. Upon inspection of this ontology, we noticed that the collection of classes and properties were missing some critical clinical elements such as smoking history, CTCAE v5 toxicity scores, diagnostic procedures such as Gleason scores, prostate-specific antigen (PSA) levels, patient reported outcome measures, Karnofsky performance status (KPS) scales, and radiation treatment modality. We utilized the ontology editor tool Protégé [26] for adding these key classes and properties in the updated ontology file. We reused entries from other published ontologies such as the National Cancer Institute's NCI Thesaurus [27], International Classification of Diseases version 10 (ICD-10) [28], and Dbpedia [29] ontologies. We added 216 classes (categories defined in Table 1) with 19 predicate elements to the ROO. With over 100,000 terms, the NCI Thesaurus includes wide coverage of cancer terms, as well as mapping with external terminologies. The NCI Thesaurus is a product of NCI Enterprise Vocabulary Services (EVS), and its vocabularies consists of public information on cancer, definitions, synonyms, and other information on almost 10,000 cancers and related diseases, 17,000 single agents and related substances, as well as other topics that are associated with cancer. The list of high-level data categories, elements, and codes that are utilized in our work are included in the appendix (Appendix A2).
| ||||||||||||||||||||||||
To use and validate the defined ontology, we mapped our data housed in the clinical data warehouse relational database with the concepts and relationships listed in the ontology. This mapping process linked each component (e.g., column headers, values) of the SQL relational database to its corresponding clinical concept (e.g., classes, relationships, and properties) in the ontology. To perform the mapping, the SQL database tables are analyzed and matched with the relevant concepts and properties in the ontology. This can be achieved by identifying the appropriate classes and relationships that best represent the data elements from the SQL relational database. For example, if the SQL relational table provides information about a patient's smoking history, the mapping process would identify the corresponding class or property in the ontology that represents smoking history.
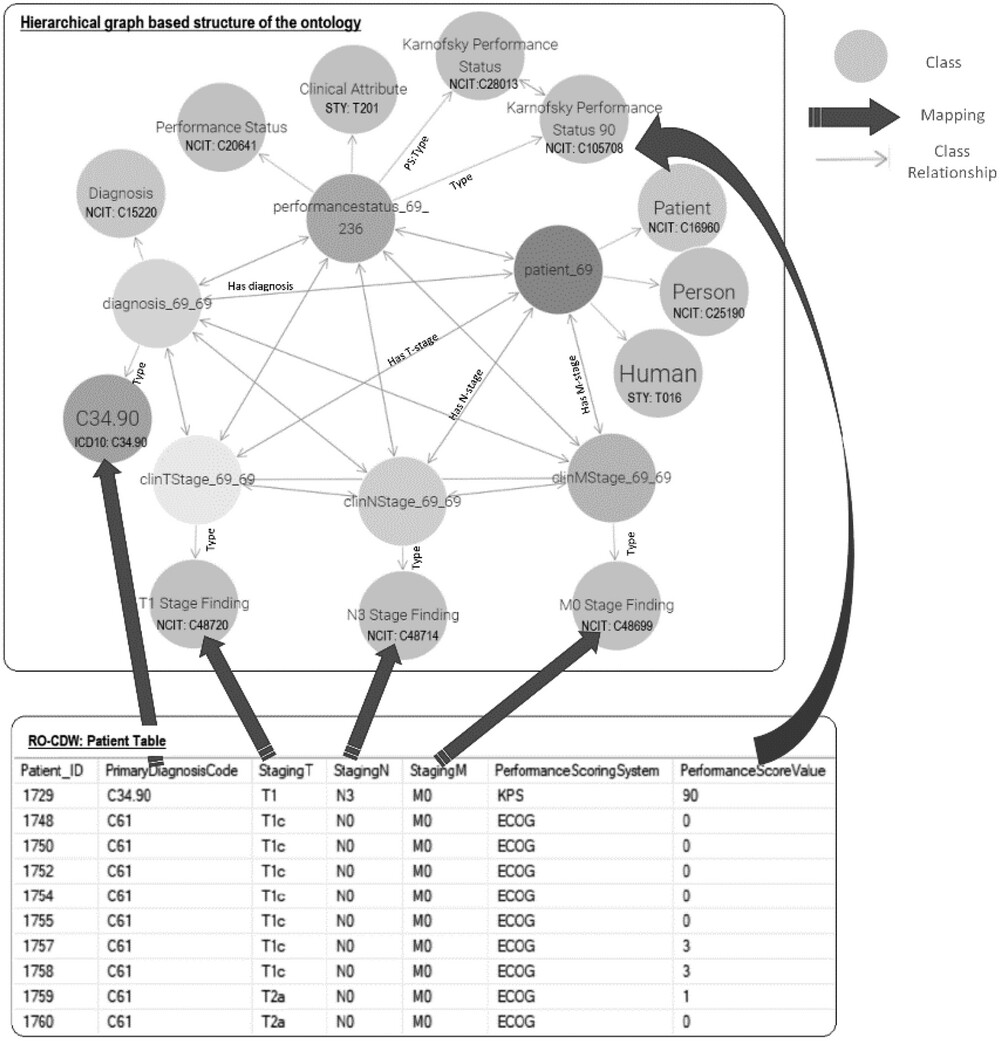
A correspondence between the table columns in the relational database and ontology entities was established using the D2RQ mapping script. An example of this mapping script is shown in Figure 2. With the use of the D2RQ mapping script, individual table columns in relational database schema were mapped to RDF ontology-based codes. This mapping script is executed by the D2RQ platform that connects to the SQL database, reads the schema, performs the mapping, and generates the output file in turtle syntax. Each SQL table column name is mapped to its corresponding class using the d2rq:ClassMap command. These classes are also mapped to existing ontology-based concept codes such as NCIT:C48720 for T1 staging. In order to define the relationships between two classes, the d2rq:refersToClassMap command is used. The properties of the different classes are defined using the d2rq:PropertyBridge command.
Unique resource identifiers (URIs) are used for each entity for enabling the data to be machine-readable and for linking with other RDF databases. The mapping process is specific to the structure and content of the ontology being used, in this case, ROO. It relies on the defined classes, properties, and relationships within the ontology to establish the mapping between the SQL tables input data and the ontology terminology. While the mapping process is specific to the published ontology, it can potentially be generalized to other clinics or healthcare settings that utilize similar ontologies. The generalizability depends on the extent of similarity and overlap between the ontology being used and the terminologies and concepts employed in other clinics. If the ontologies share similar structures and cover similar clinical domains, the mapping process can be applied with appropriate adjustments to accommodate the specific terminologies and concepts used in the target clinic.
|
Importing data in knowledge based graph-based database
The output file from the D2RQ mapping step is in Terse RDF Triple Language (turtle) syntax. This syntax is used for representing data in the semantic triples, which comprise a subject, predicate, and object. Each item in the triple is expressed as a Web URI. In order to search data from such formatted datasets, the dataset is imported in RDF knowledge graph databases. An RDF database, also called a Triplestore, is a type of graph database that stores RDF triples. The knowledge on the subject is represented in these triple formats consisting of subject, predicate, and object. An RDF knowledge graph can also be defined as labeled multi-diagraphs, which consist of a set of nodes which could be URIs or literals containing raw data, and the edges between these nodes represent the predicates. [30] The language used to reach data is called SPARQL—Query Language for RDF. It contains ontologies that are schema models of the database. Although SPARQL adopts various structures of SQL query language, SPARQL uses navigational-based approaches on the RDG graphs to query the data, which is quite different than the table-join-based storage and retrieval methods adopted in relational databases. In our work, we utilized the Ontotext GraphDB software [31] as our RDF store and SPARQL endpoint.
Ontology keyword-based searching tool
It is common practice amongst healthcare providers to use different medical terms to refer to the same clinical concept. For example, if the user is searching for patient records that had a “heart attack,” then besides this text word search, they should also search for synonym concepts such as “myocardial infarction,” “acute coronary syndrome,” and so on. Ontologies such as NCI Thesaurus have listed synonym terms for each clinical concept. To provide an effective method to search the graph database, we built an ontology-based keyword search engine that utilizes the synonym-based term-matching methods. Another advantage of using ontology-based term searching is realized by using the class parent-children relationships. Ontologies are hierarchical in nature, with the terms in the hierarchy often forming a directed acyclic graph (DAG). For example, if we are searching for patients in our database with clinical stage T1, the matching patient list will only comprise patients that have T1 stage NCI Thesaurus code (NCIT: C48720) in the graph database. These matching patients will not return any patients with T1a, T1b, and T1c sub-categories that are children of the parent T1 staging class. We built this search engine where we can search on any clinical term and its matching patient records based on both parent and children classes, which are abstracted.
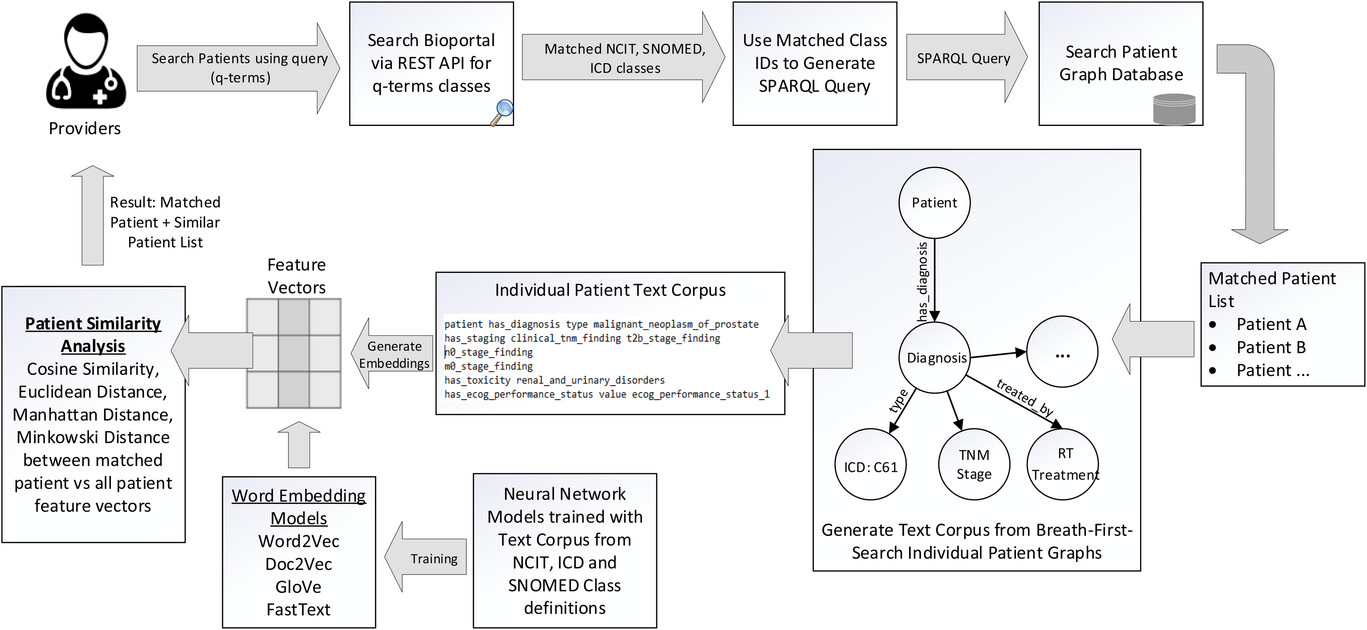
The method that is used in this search engine is as follows. When the user wants to use the ontology to query the graph-based medical records, the only input necessary is the clinical query terms (q-terms) and an indication of whether the synonyms should also be considered while retrieving the patient records. The user has the option to specify the multiple levels of child class search and parent classes to be included in the search parameters. The software will then connect to the Bioportal database via REST API and perform the search to gather the matching classes for the q-terms and the options specified in the program. Using the list of matching classes, a SPARQL-based query is generated and executed with our patient graph database and matching patient list, and the q-term based clinical attributes are returned to the user.
In order to find patients that have not the same but similar attributes based on the search parameters, we have designed a patient similarity search method. The method employed to identify similar patients based on matching knowledge graph attributes involves the creation of a text corpus by performing breadth-first search (BFS) random walks on each patient's individual knowledge graph. This process allows us to explore the graph structure and extract the necessary information for analysis. Within each patient's knowledge graph, approximately 18−25 categorical features were extracted in the text corpus. It is important to note that the number of features extracted from each patient may vary, as it depends on the available data and the complexity of the patient's profile. These features included the diagnosis; tumour, node, metastasis (TNM) staging; histology; smoking status; performance status; pathology details; radiation treatment modality; technique; and toxicity grades.
This text corpus is then used to create word embeddings that can be used later to search for similar patients based on similarity and distance metrics. We utilized four vector embedding models, namely Word2Vec [32], Doc2Vec [33], GloVe [34], and FastText [35] to train and generate vector embeddings. The output of word embedding models are vectors, one for each word in the training dictionary, that effectively capture relationships between words. The architecture of these word embedding models is based on a single hidden layer neural network. The description of these models is provided in Appendix A1.
The text corpus used for training is obtained from the Bioportal website, which encompasses NCIT, ICD, and SNOMED codes, as well as class definition text, synonyms, hyponyms terms, parent classes, and sibling classes. We scraped 139,916 classes from the Bioportal website using API calls and used this dataset to train our word embedding models. By incorporating this diverse and comprehensive dataset, we aimed to capture the semantic relationships and contextual information relevant to the medical domain. The training process involved iterating over the training dataset for a total of 100 epochs using CPU hardware. During training, the models learned the underlying patterns and semantic associations within the text corpus, enabling them to generate meaningful vector representations for individual words, phrases, or documents. Once the models were trained, we utilized them to generate vector embeddings for the individual patient text corpus that we had previously obtained. These embeddings served as numerical representations of the patient data, capturing the semantic and contextual information contained within the patient-specific text corpus. The Cosine similarity, Euclidean distance, Manhattan distance, and Minkowski distance metrics are employed to measure the distance between the matched patients and all patient feature vectors.
Figure 3 shows the design architecture of the software system. The main purpose of this search engine is to provide the users with a simple interface to search the patient records.
|
Results
Mapping data to the ontology
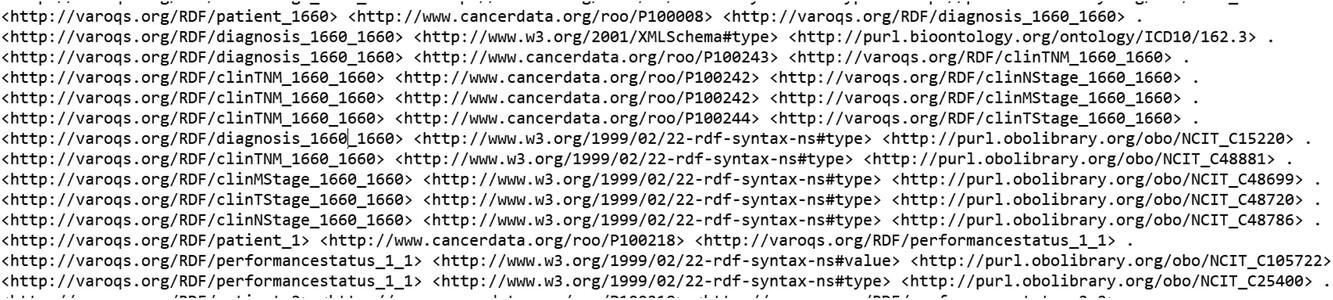
With the aim of testing out the data pipeline and infrastructure, we used our clinical database that has 1,660 patient clinical and dosimetry records. These records are from patients treated with radiotherapy for prostate cancer, non-small cell lung cancer, and small cell lung cancer disease. There are 35,303 clinical and 12,565 DVH based data elements that are stored in our RO-CDW database for these patients. All these data elements were mapped to the ontology using the D2RQ mapping language, resulting in 504,180 RDF tuples. In addition to the raw data, these tuples also defined the interrelationships amongst various defined classes in the dataset. An example of the output RDF tuple file is shown in Figure 4, displaying the patient record relationship with diagnosis, TNM staging, etc. All the entities and predicates in the output RDF file have a URI, which is resolvable as a link for the computer program or human to gather more data on the entities or class. For example, the RDF viewer would be able to resolve the address http://purl.obolibrary.org/obo/NCIT_48720 to gather details on the T-stage such as concept definitions, synonym, relationship with other concepts and classes, etc.
We were able to achieve a mapping completeness of 94.19% between the records in our clinical database and RDF tuples. During the validation process, we identified several ambiguities or inconsistencies in the data housed in the relational database, such as indication of use of ECOG instrument for performance status evaluation but missing values for ECOG performance status score, record of T stage but nodal and metastatic stage missing, and delivered number fractions missing with the prescribed dose information. To maintain data integrity and accuracy, the D2RQ mapping script was designed to drop these values due to missing or incomplete data or ambiguous information. Additionally, the validation process thoroughly examined the interrelationships among the defined classes in the dataset. We verified that the relationships and associations between entities in the RDF tuples accurately reflected the relationships present in the original clinical data. Any discrepancies or inconsistencies found during this analysis were identified and addressed to ensure the fidelity of the mapped data. To evaluate the accuracy of the mapping process, we conducted manual spot checks on a subset of the RDF tuples. This involved randomly selecting samples of RDF tuples and comparing the mapped values to the original data sources. Through these spot checks, we ensured that the mapping process accurately represented and preserved the information from the clinical and dosimetry data during the transformation into RDF tuples. Overall, the validation process provided assurance that the pipeline effectively transformed the clinical and dosimetry data stored in the RO-CDW database into RDF tuples while preserving the integrity, accuracy, and relationships of the original data.
|
Visualization of data in ontology-based graphical format
Visualizations on ontologies play a key role for users to understand the structure of the data and work with the dataset and its applications. This has an appealing potential when it comes to exploring or verifying complex and large collections of data such as ontologies. We utilized the Allegrograph Gruff toolkit [36] that enables users to create visual knowledge graphs that display data relationships in a neat graphical user interface (GUI). The Gruff toolkit uses simple SPARQL queries to gather the data for rendering the graph with nodes and edges. These visualizations are useful because they increase the users’ understanding of data by instantly illustrating relevant relationships amongst class and concepts, hidden patterns, and data significance to outcomes. An example of the graph-based visualization for a prostate and non-small cell lung cancer patient is shown in Figures 5 and 6. Here all the nodes stand for concepts and classes, and the edges represent relationships between these concepts. All the nodes in the graph have URIs that are resolvable as a web link for the computer program or human to gather more data on the entities or classes. The color of the nodes in the graph visualization are based on the node type, and there are inherent properties of each node that include the unique system code (e.g., NCIT code or ICD code, etc.), synonyms terms, definitions, value type (e.g., string, integer, floating point number, etc.). The edges connecting the nodes are defined as properties and stored as predicates in the ontology data file. The use of these predicates enables the computer program to effectively find the queried nodes and their interrelationships. Each of these properties are defined with URIs that are available for gathering more detailed information on the relationship definitions. The left panel in Figures 5 and 6 shows various property types or relationship types that connect the nodes in the graph. Using SPARQL language and Gruff visualization tools, users can query the data without having any prior knowledge of the relational database structure or schema, since these SPARQL queries are based on universal publish classes defined in the NCI Thesaurus, Units Ontology, and ICD-10 ontologies.
|
|
Finally, these SPARQL queries can be used with commonly available programming languages like Python and R via REST APIs. We also verified data from the SPARQL queries and the SQL queries from the CDW database for accuracy of the mapping. Our analysis found no difference in the resultant data from the two query techniques. The main advantage of using the SPARQL method is that the data can be queried without any prior knowledge of the original data structure based on the universal concepts defined in the ontology. Also, the data from multiple sources can be seamlessly integrated in the RDF graph database without the use of complex data matching techniques and schema modifications, which is currently required with relational databases. This is only possible if all the data stored in the RDF graph database refers to published codes from the commonly used ontologies.
Searching the data using ontology-based keywords
For effective searching of discrete data from the RDF graph database, we built an ontology-based keyword searching web tool. The public website for this tool is https://hinge-ontology-search.anvil.app. Here we are able to search the database based on keywords (q-terms). The tool is connected to the Bioportal via REST API and finds the matching classes or concepts and renders the results including the class name, NCIT code, and definitions. We specifically used the NCI Thesaurus ontology for our query which is 112 MB in size and contains approximately 64,000 terms. The search tool can find the classes based on synonym term queries where it matches the q-terms with the listed synonym terms in the classes (Figure 7a). The tool has features to search the child and parent classes on the matching q-term classes. A screenshot of the web tool with the child class search is shown in Figure 7b. The user can also specify the level of search ,which indicates if the returned classes should include classes of children of children. In the example in Figure 9b, we are showing the q-term used for searching “fatigue” while including the child classes up to one level, and the return classes included the fatigue based CTCAE class and the grade 1, 2, 3 fatigue classes. Once all the classes used for searching are found by the tool, it searches the RDF graph database for matching patient cases with these classes. The matching patient list, including the found class in the patient's graph, is displayed to the user. This tool is convenient for end users to abstract cohorts of patients that have particular classes or concepts in their records without the user learning and implementing the complex SPARQL query language. Based on our evaluation, we found that the average time taken to obtain results is less than five seconds per q-term if there are less than five child classes in the query. The maximum time taken is 11 seconds for a q-term that had 16 child classes.
|
For evaluating the patient similarity-based work embedding models, we evaluated the quality of the feature embedding-based vectors produced by using the technique called t-Distributed Stochastic Neighbor Embedding (t-SNE) and cluster analysis with a predetermined number of clusters set to five based on the diagnosis groups for our patient cohort. Our main objective is to determine the similarity between patient data that are in the same cluster based on their corresponding diagnosis groups. This method can reveal the local and global features encoded by the feature vectors and thus can be used to visualize clusters within the data. We applied t-SNE to all 1,660 patient feature-based vectors produced via the four word embedding models. The t-SNE plot is shown in Figure 8; it shows that the disease data points can be grouped into five clusters with varying degrees of separability and overlap. The analysis of patient similarity using different embedding models revealed interesting patterns. The Word2Vec model showed the highest mean cosine similarity of 0.902, indicating a relatively higher level of similarity among patient embeddings within the five diagnosis groups. In contrast, the Doc2Vec model exhibited a lower mean cosine similarity of 0.637. The GloVe model demonstrated a moderate mean cosine similarity of 0.801, while the FastText model achieved a similar level of 0.855. Regarding distance metrics, the GloVe model displayed lower mean Euclidean and Manhattan distances, suggesting that patient embeddings derived from this model were more compact and closer in proximity. Conversely, the Doc2Vec, Word2Vec, and FastText models yielded higher mean distances, indicating greater variation and dispersion among the patient embeddings. These findings provide valuable insights into the performance of different embedding models for capturing patient similarity, facilitating improved understanding and decision-making in the clinical domain.
|
Discussion
Abbreviations, acronyms, and initialisms
Acknowledgements
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, though grammar and word usage was substantially updated for improved readability. In some cases important information was missing from the references, and that information was added.