File:Fig8 Garza BMCBioinformatics2016 17.gif
Fig8_Garza_BMCBioinformatics2016_17.gif (566 × 415 pixels, file size: 63 KB, MIME type: image/gif)
Summary
| Description |
Fig. 8 Overview of how CTDs interact with programming languages and workflow systems. CTDs can be generated by CTD-enabled tools (e.g., BALL, OpenMS, SeqAn) or via CTDopts. Once a tool is CTD-enabled, it can be imported into the KNIME Analytics Platform or Galaxy. We have also developed converters that can import KNIME Analytics Platform, Galaxy workflows into gUSE to take advantage of HPC resources and DCIs. |
|---|---|
| Source |
de la Garza, L.; Veit, J.; Szolek, A.; Röttig, M.; Aiche, S.; Gesing, S.; Reinert, K.; Kohlbacher, O. (2016). "From the desktop to the grid: Scalable bioinformatics via workflow conversion". BMC Bioinformatics 17: 127. doi:10.1186/s12859-016-0978-9. PMC PMC4788856. PMID 26968893. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4788856. |
| Date |
2016 |
| Author |
de la Garza, L.; Veit, J.; Szolek, A.; Röttig, M.; Aiche, S.; Gesing, S.; Reinert, K.; Kohlbacher, O. |
| Permission (Reusing this file) |
|
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution 4.0 License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 16:14, 14 June 2016 | 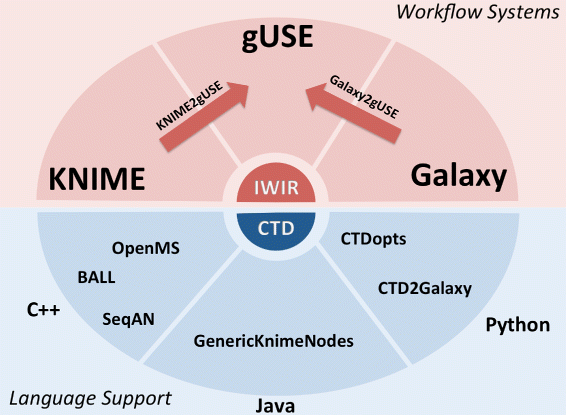 | 566 × 415 (63 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file:









