Journal:A robust, format-agnostic scientific data transfer framework
| Full article title | A robust, format-agnostic scientific data transfer framework |
|---|---|
| Journal | Data Science Journal |
| Author(s) | Hester, James |
| Author affiliation(s) | Australian Nuclear Science and Technology Organisation |
| Primary contact | Email: jxh at ansto dot gov dot au |
| Year published | 2016 |
| Volume and issue | 15 |
| Page(s) | 12 |
| DOI | 10.5334/dsj-2016-012 |
| ISSN | 1683-1470 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://datascience.codata.org/articles/10.5334/dsj-2016-012/ |
| Download | http://datascience.codata.org/articles/10.5334/dsj-2016-012/galley/605/download/ (PDF) |
Abstract
The olog approach of Spivak and Kent[1] is applied to the practical development of data transfer frameworks, yielding simple rules for construction and assessment of data transfer standards. The simplicity, extensibility and modularity of such descriptions allows discipline experts unfamiliar with complex ontological constructs or toolsets to synthesize multiple pre-existing standards, potentially including a variety of file formats, into a single overarching ontology. These ontologies nevertheless capture all scientifically-relevant prior knowledge, and when expressed in machine-readable form are sufficiently expressive to mediate translation between legacy and modern data formats. A format-independent programming interface informed by this ontology consists of six functions, of which only two handle data. Demonstration software implementing this interface is used to translate between two common diffraction image formats using such an ontology in place of an intermediate format.
Keywords: metadata, ontology, knowledge representation, data formats
Introduction
For most of scientific history, results and data were communicated using words and numbers on paper, with correct interpretation of this information reliant on the informal standards created by scholarly reference works, linguistic background, and educational traditions. Modern scientists increasingly rely on computers to perform such data transfer, and in this context the sender and receiver agree on the meaning of the data via a specification as interpreted by authors of the sending and receiving software. Recent calls to preserve raw data[2][3] and a growing awareness of a need to manage the explosion in the variety and quantity of data produced by modern large-scale experimental facilities (big data) have led to an increase in the number and coverage of these data transfer standards. Overlap in the areas of knowledge covered by each standard is increasingly common, either because the newer standards aim to replace older ad hoc or de facto standards, or because of natural expansion into the territory of ontologically “neighboring” standards. One example of such overlap is found in single-crystal diffraction: the newer NeXus standard for raw data[4] partly covers the same ontological space as the older imgCIF standard[5], and both aim to replace the multiplicity of ad hoc standards for diffraction images.
Authors of scientific software faced with multiple standards generally write custom input or output modules for each standard. For example, the HKL Research, Inc. suite of diffraction image processing programs accepts over 300 different formats.[6] In such software, broadly useful information on equivalences and transformations is crystallized in code that is specific to a programming language and software environment and is therefore difficult for other authors faced with the same problems to reuse, even if code is freely available. Such uniform processing and merging of disparate standards has been extensively studied by the knowledge representation community: it is one outcome of "ontological alignment" or "ontological mapping," which has been the subject of hundreds of publications over the last decade.[7] Despite the availability of ontological mapping tools, Otero-Cerdeira, Rodríguez-Martínez, & Gómez-Rodríguez note that relatively few ontology matching systems are put to practical use (see their section 4.5). One barrier to adoption is likely to be the need for the discipline experts driving standards development to learn ontological concepts and terminology in order to evaluate and use ontological tools: the effort required to master these tools may not be judged to yield commensurate benefits in situations where communities have historically been able to transfer data reliably without such formal approaches. Introduction of ontological ideas into data transfer would therefore stand more chance of success if those ideas are simple to understand and implement, as well as offering tangible benefits over the status quo. Indeed one of the challenges noted by Otero-Cerdeira et al. is to "define good tools that are easy to use for non-experts."
Much of the research listed by Otero-Cerdeira et al. has understandably been predicated on reducing human involvement in the mapping process, although expert human intervention is still currently required. In contrast to the thousands of terms found in ontologies tackled by ontological mapping projects, data files in the experimental sciences usually contain information relating to a few dozen well-defined scientific concepts, and so manual handling of ontologies is feasible. The present paper therefore adopts the practical position that, if involvement of discipline experts is unavoidable, then the method of representing the ontology should be as accessible as possible to those experts. An easily-applied framework for scientist-driven formalization, development and assessment of data transfer standards is presented, aimed at minimizing the complexity of the task, while promoting interoperability and minimizing duplication of programmer and domain expert effort.
After describing the framework in the next section, we demonstrate the utility of these concepts by discussing schemes for standards development and later semiautomatic data file translation.
A conceptual framework for data file standards
The framework described here covers systems for automated transfer and manipulation of scientific data. In other words, following creation of the reading and writing software in consultation with the data standard, no further human intervention is necessary in order to automatically create, ingest, and perform calculations on data from standards-conformant data files. Note that simple transfer of information found in the data file to a human reader — for example, presentation of text or graphics — is of minor significance in this context, as such operations, while useful, do not require any interpretation of the data by the computer and are in essence identical to traditional paper-based transfer of information from writer to reader.
Terminology used in this paper is defined in Table 1. The process of scientific data transfer is described using these terms as follows: in consultation with the ontology, authors of file output software determine the required or possible list of datanames for their particular application, then correlate concepts handled by their code to these datanames, arranging for the appropriate values to be linked to the datanames within the output data format according to the specifications within the format adapter. A file in this format is then transferred or archived. At some point, software written in consultation with the same format adapter and ontology extracts datavalues from the file and processes them correctly.
| ||||||||||||||||||||
Following Shvaiko & Euzenat[8], the word "ontology" as used in this paper refers to a system of interrelated terms and their meanings, regardless of the way in which those meanings are represented or described. Under this definition, Table 1 is itself an ontology for use solely by the human reader in understanding the present paper. An ontology may be encoded using a language such as OWL[9] to produce a human-and machine-readable document allowing some level of machine verification, deduction and manipulation.
This paper makes frequent reference to two established data transfer standards in the area of experimental science: the Crystallographic Information Framework (CIF)[10] and the NeXus standard.[11]
Constructing the ontology
In general, a complete data transfer ontology for some field would include all of the distinct concepts and relationships used by scientific software authors in the process of constructing software, including scientific, programming, and format-specific terminology. A clear dividing line may be drawn between the scientific components of the ontology and the remainder, by relying on the assertion that scientific concepts and their relationships are dictated by the real world, not by the particular arrangement in which the data appear; that is, a scientific ontology may be completely specified independent of a particular format.
Furthermore, the scheme presented below assumes that the scientific knowledge informing the ontology is already shared by the software authors implementing the standard. These software authors are one of the main consumers of the ontology, so we do not require the level of machine-readability offered by ontology description languages such as OWL; rather we seek the minimum level of sophistication necessary to describe to a human the correct interpretation of the data, while at the same time including properties that allow coherent expansion and curation of the ontology. Such ontologies should be maximally accessible to experts in the scientific field who are not necessarily programmers or familiar with ontological constructs, in order to allow broad-based contribution and review.
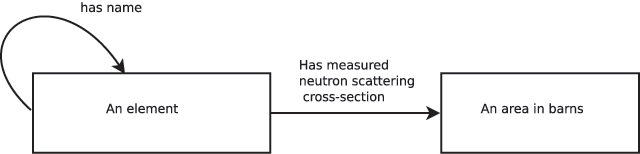
A suitably simple but powerful system for expressing ontologies has been presented by Spivak & Kent[1], who propose using category-theoretic box and arrow diagrams which they call ologs (from "ontology logs"). A concept in an ontology is drawn as an arrow ("aspect") between boxes ("types"): the arrow denotes a mapping between elements in the sets represented by the boxes. A simple ontology written using this approach is shown in Figure 1, which might be used to describe a data file containing the values of neutron cross-section. This olog shows that the concept "measured neutron scattering cross-section" maps every atomic element to a value in barns. We can therefore specify "measured neutron scattering cross-section" as a (domain, function, codomain)[a] triple of ({element names},‘cross-section measurement’,{(r,“barns”) : r ∈ ℝ}). Each of the datanames in our ontology is associated with such a triple, so that the values that the dataname takes are the results of applying the associated function to each of the elements of the domain. Given this formulation of an ontology, it follows that the scientifically useful content of a datafile consists solely of the values taken by the datanames in their codomains, and the matching domain values. In other words, a datafile documents an instance of the olog.
|
Such a functional formulation of a scientific area is generally trivially available. For example, when the result of applying the function might not be known for all elements of the domain, we can augment the co-domain with a null value.[12] Where the function requires several parameters in order to determine uniquely an element of the co-domain, we simply define the domain to be a tuple of the appropriate length. Where a concept would map a single domain element to multiple values and we cannot invert the direction of the relation, the co-domain can be defined to consist of tuples. The identity function applied to a type also creates a dataname, which identifies objects of that type: for example "element name," "measurement number" or "detector bank identifier."
It is useful to visualize the collection of datanames that are functions of the same domain as forming a table; a row in this table describes the mappings of a single value from the domain into the co-domains of the other datanames. Indeed, an olog is trivially transformable to a relational database schema[13], in which case datanames are equivalent to database table column names. Note that this close link to relational databases in no way requires us to use a relational database format for data transfer, although a relational database may serve as a useful baseline against which to compare the chosen data format.
As the above description is potentially too technical to meet our stated aims of broad accessibility, we may instead describe an olog as a collection of definitions for datanames, where each definition meets the following requirements:
- a description
- a description of the concept sufficient for a human reader to unambiguously reconstruct a mapping between the one or more related items and the value type. Every unique collection of values for related items must map to a single value of value type.
- value type
- (the codomain of the function). While the particular discipline will often determine the types of values, a versatile and generally suitable set of core value types might consist of: numeric types (integer,real,complex), finite sets of alternatives (numerical or text), and homogeneous compound structures composed of such values (arrays, matrices). Units should be specified where relevant. Free-form text is useful when constructing an opaque identifier (e.g., sample name), although in other contexts it will not be machine interpretable.
- related items
- dataname(s) that this item maps from (collectively forming the domain of the function).[b] This includes all names which participate in determining the behavior of the mapping, for example, it includes all parameters influencing a model calculation. If it is not possible to identify a unique value for the dataname given only the specific values of the datanames in this list, then the related items list is incomplete. In order to completely insulate users of the specification from expansions in the ontology, once defined this list of related items cannot change (see the next section).
As expected, an ontology constructed according to the above prescription is completely decoupled from the file format. Any discipline specialist able to provide the above information can contribute to the ontology regardless of programming skill, thus broadening the base of contributors.
Although our objective is to provide an ontology that is easy for non-programmers to contribute to, there is some advantage to producing a computer-readable version of the ontology, which might contain additional validation information, e.g., range limits on values that are otherwise only provided in human-readable form (perhaps implicitly) in a plain-text ontology. An olog could, for example, be encoded in the popular machine-readable OWL formalism by specifying an OWL "class" for each olog "type" and creating OWL "functional object properties" for each olog aspect (that is, a dataname is an OWL functional object property linking two OWL classes). OWL terms for restricting value ranges would be used to further describe each OWL class. In general, a restricted and well-chosen vocabulary for the machine-readable ontology will also prompt the ontology author to include useful information that might otherwise be assumed (for example, restricting a range to positive non-zero numbers). An extended example later shows how information presented in a machine-readable ontology can be leveraged for automatic format conversion.
Note that a definition may also stipulate default values. As a reasonably large ontology may contain several alternative routes for reaching a given value (indeed, the notion of path equivalence is a defining feature of ologs and mathematical categories), some care should be taken when stating default values to ensure that they match up with those stated for dependent items.
Expanding ontologies
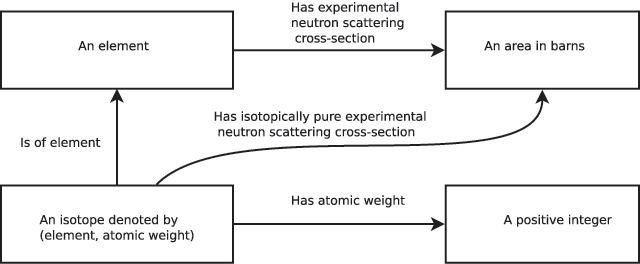
To be useful in the long term, a scientific ontology must be able to gracefully incorporate new scientific discoveries and concepts. In our simple ontological scheme, such expansion is accomplished by addition of new types and aspects. Addition of a new aspect between existing types is clearly not problematic. Addition of a new type (accompanied by an identifier dataname) may, however, include the assertion that a previously-defined dataname is additionally or instead dependent on the newly-defined type. This would potentially impact on all software produced prior to the change, as calculations in older software would not take into account the effect of the new type. As such an impact is almost always undesirable, a fundamental rule should be adopted that the list of dependencies of a dataname may never change. Instead, in this scenario a duplicate of the original dataname should be introduced that includes the new dependency, with the addition of any appropriate functional mappings involving the new and old datanames. For example, consider our simple ontology of Figure 1 following the "discovery" that neutron scattering cross-section depends both on element and atomic weight (Figure 2). Rather than redefining our "experimental neutron scattering cross-section" domain, which consisted solely of the element name, the new concept of "an isotope" (that is, element combined with atomic weight to form a pair) is introduced. A new dataname "isotopically pure experimental neutron scattering cross-section"[c] maps isotope to cross-sectional area. Note that while it is inadvisable to rename or change the dependencies of the old dataname, the descriptive part of the old dataname’s definition can certainly be updated to explain that this dataname refers to the scattering cross-section for natural isotope abundance (if that is indeed what was originally measured).
|
Managing large ontologies
Any reasonably large ontology will benefit from human-friendly organizational tools. The following three suggestions may be useful:
- Adopting a naming convention <type><separator><name> (e.g., "measurement:intensity"), such that all datanames mapping from the same type have the same first component in the name. This makes it easier to find datanames, and is also useful for machine-readable ontologies.
- Defining type "groups," where all types of a given "group" are guaranteed to have the same minimum set of datanames mapping from them.[d] This is most useful in translation scenarios where the ontology writers cannot control the concepts found in foreign data files. For example, a legacy standard may separate "sample manipulation axis" and "detector axis"; an alternative ontology might simply define an "axis" type. A proper description of the contents of the legacy data file requires defining datanames for the two types of axis, in which case assigning an axis "group" to all "axis," "sample manipulation axis" and "detector axis" types saves space, redefining all of the common attributes of an axis. Note that we should not use the format adapter to perform such legacy manipulations as, by design, we wish it to contain no scientific knowledge (see below).
- Separating ontologies into a "core" or "general" ontology, and subsidiary ontologies that define transformations of items in the core ontology. This allows datanames created for legacy data files whose values are subsets of values of datanames in the core ontology to be separated out, reducing clutter and repetition.
Units
The ontology must specify units in order to avoid ambiguity. Not specifying units is equivalent to allowing the elements of a codomain to be represented as a pair: (real value, units), where units can have different values (e.g., "mm" or "cm"), breaking our basic rule that each element of the domain maps to a single element of the co-domain: for example, "1 cm" is the same as "0.01 m" and therefore any value that maps to "(1, cm)" maps also to "(0.01, m)". The units of measurement are therefore a characteristic of the type as a whole, that is, units should always be specified for olog types where units are defined. Where multiple units for a single concept are in common use and the community cannot agree on a single choice, separate types must be defined together with separate datanames mapping to those types.
Uncertainties
Standard uncertainties (su) are sometimes included as part of the data value (for example, in the CIF standard), resulting in a data value that is a pair of numbers. This may be convenient in situations where functions of the data value need to propagate errors. Alternatively, the su can easily be assigned a separate dataname as the mapping from measurement to su is unambiguous. In such a case, when the (measurement, su) pair is required, an additional dataname corresponding to this is defined.
The dataname list
A data bundle contains the values for some set of datanames. The list of datanames in the bundle, together with the ontology, completely specifies the semantic content of the transferred data. A data bundle may be further characterized in practice by indicating which datanames (if any) must be unvarying within the bundle. A single value for a constant-valued dataname can be supplied in the file, and then datanames that are dependent on such constant-valued datanames do not need to explicitly link their values with the corresponding constant-valued dataname value, leading to simplification in data file structure. In the following, for simplicity "data file" and "data bundle" are used interchangeably, although in general a data file may include multiple data bundles; in the CIF and NeXus contexts the bundles inside the data files are called "data blocks" and "entries" respectively.
The specification of such dataname lists for a bundle may evolve over time in three ways: (i) addition of single-valued datanames; (ii) addition of one or more multiple-valued datanames; (iii) change of a single-valued dataname to a multi-valued dataname. The considerable difficulty involved in distributing updated software in a timely manner requires that there is no impact of such evolution on already-existing software, that is, if the dataname list contained within a file produced according to the standard is updated, that file must remain compatible with software that reads older files. The first two cases meet this requirement given the stipulation on ontology expansion (discussed in a previous section). If, however, a single-valued dataname becomes multi-valued as in case (iii), software that expects a single value will almost certainly fail. This failure is usually due to the fact that, in addition to a dataname becoming multi-valued, a series of other values for dependent datanames must now be connected to the appropriate value of the newly multi-valued dataname using some format-specific mechanism. Note that a simple algorithmic transformation exists to transform the new-style files to old-style files: for every value of the newly multi-valued dataname, a data bundle with this single value for the dataname is produced. This means that file reading software could, in theory, automatically handle even this type of evolution in the dataname list. Software written in this way appears to be rare in practice however, presumably because moving to a multi-valued dataname will often imply a different type of analysis and therefore significantly different or upgraded software, which is then written to handle older-style files internally as a special case.
To successfully transfer data files automatically, authors of both file creation and file reading software must agree on the contents of the dataname list to the extent that the file reading software requires a subset of the list placed in the file by the file creation software. One simple but initially time-consuming strategy to achieve such coordination is for the file authoring software to place into output files as many data items from the ontology as are available; if this is insufficient for a given application, the problem lies with the experiment or ontology rather than with the authoring software. Another form of coordination arises based on the use case: a sufficiently well-understood use case will imply a set of concepts from the ontology on which both reader and writer will independently agree. For example, the use case "display a single-crystal atomic structure" immediately implies a list of crystal structure datanames, which can be encoded into an output file by structural databases and then read and used successfully by structural display programs, without any direct coordination between the parties. Similar scenarios determine the contents of dataname lists found in ad-hoc standards based around input and output files for particular software packages.
Long-term archiving is an important special case, as the specific use to which the data retrieved from the archive will be put is not known. Archives therefore aim to include as much information as possible; database deposition software acts to enforce the corresponding dataname list (even if this requirement is not phrased in terms of datanames), and the feedback loop via database depositors to the authors of their software eventually creates a stable dataname list. The IUCr "CheckCIF" program[14]) works in a similar way to enforce a dataname list for publication of crystallographic results.
Choosing a file format
Given that our data is simply an instance of our olog, a file format at minimum must allow (i) association of datanames with sets of co-domain values, and (ii) matching co-domain values to the domain values for each dataname. Any other capabilities provided by the format are not ontologically relevant, but may have important practical implications, for example efficient transfer or storage, or suitability for long-term archiving. Allowing for expansion and variety in dataname lists from the outset is important: as discussed above, this means allowing for the possible addition of new single- and multiple-valued datanames, where the multiple-valued datanames may be independent of the already-included datanames (in database terms, a new table might be required).[e] If such a format is not chosen initially, the alternative is to move to a new format when faced with a use case requiring, e.g., more columns than the format allows. Although integration of a new format into a data transfer specification is dramatically simplified by the present approach, the impact of adopting a new format on the distributed software ecosystem that would have developed around the previous format would still be considerable, meaning that the choice of a flexible format capable of containing multiple independently varying datanames (i.e., multiple tables in database terms) is strongly advisable when designing any new standard.
Certain types of value found in the ontology may not be easily or efficiently represented by a given file format: the format may not provide a structure that can efficiently represent a matrix, for example. Just as for dataname location issues, the ontology is impervious to such data representation issues; rather, it is up to the authors of the transfer specification to choose a format which is the best fit to the dataname list which their use case requires.[f]
Elements of a format adapter
The format adapter describes how the values for the datanames in the dataname list are encapsulated in a data format. The format adapter provides the following information:
- format
- the file format used
- location in format
- how values corresponding to each name in the dataname list are located in the chosen format
- value representation
- how each type of value is represented in this format (if at all), including how missing values are reported
It is often the case that a standard third-party format (e.g., XML or HDF5) offers choices for the way in which multiple datavalues can be encapsulated or a particular value is expressed. In such cases, the format adapter builds on the data format specification to delimit which of the available options have been chosen. For example, the NeXus standard uses custom HDF5[15] attributes to locate datanames, rather than logical HDF5 paths (see below). For simple formats no explicit format adapter specification may be necessary.
By design the format adapter embodies no knowledge of the scientific ontology. This stipulation ensures that scientific domain knowledge is not linked to any particular format, and is accessible to ontology authors and readers. In programming terms, code implementing a format adapter should never rely on knowledge of a dataname when performing calculations on values. Format adapters may therefore compress and expand values, but not calculate values for missing datanames from datanames that are present. Where a format adapter author requires new datanames, a supplementary ontology should be created.
Application I: Developing an ontological commons
Using pre-existing specifications to create the commons
An advantage of the current approach is that ontological information can be harvested from multiple preexisting standards and placed into an "ontological commons," which can then act as a community resource for updating old standards and creating new standards. When new standards draw on such a commons, older data files can be automatically translated to the new format as datanames in the new ontology will be a superset of those found in the older standards.
A procedure for extraction of datanames from old standards and conversion to standard form for inclusion in such a commons requires unpacking the approaches used by particular standards designers to meet practical goals such as efficiency or simplicity. While the term "dataname" is not necessarily used in specification material, the scientific content of all data formats can be simply described as a collection of values for datanames. In this context, a "dataname" is a function that maps a set of locations in the file structure to one or more values corresponding to an olog type: examples of such locations include nodes in a hierarchical tree at the same level or of the same class, a column number, a field number in each line, or a byte offset. In addition to the easily-identified single-valued datanames, any logical structure that allows multiple values to be associated with an object is a potential dataname. The following guide to creating an ontological commons includes examples of common location types of which the author is aware.
- For each standard, identify the types. The list for value type discussed earlier is likely to cover most types found in typical data formats.
- As discussed above, sets of logical locations that map to one or more values of a single type are assigned datanames. The nature of these locations is entirely determined by the particular file format. As specifications, in order to be useful, must link operations on the file with scientifically relevant information, the linkage between scientific concept and logical location is usually simple to determine. As well as these obvious location specifications, the possible structural relationships for each format used by the standard are examined, and additional datanames are created when those structural relationships are used to encapsulate information. If structural relationships are only used to create the correspondence between values of multiple-valued datanames no dataname is created. Typical examples of "structural" datanames include the order of appearance of a value being significant (so an additional dataname taking sequential values might be required) or a parent in a hierarchy being significant (so the identifier of the parent is recorded in a new dataname). On the other hand, if a structural relationship is common to all datafiles for a given standard (e.g., "detector" is always a child of "instrument"), it is unlikely to be significant as it conveys no dataset-specific information. Locations that refer solely to aspects of the format (e.g., compression type, format version) are ignored.
- The union of the sets of datanames from all the component standards results in the overall ontology. When performing the union, datanames with the same apparent meaning but different dependencies must be distinguished, and datanames may only depend on datanames that are either also defined within that ontology, or else may be assumed to take a constant, default value: a standard that includes datanames "A" and "B" could state that "A" is a dependency of "B" whereas a similar assumption can only be made for a standard that does not include the concept "A" if it is possible to deduce that concept "A" has a constant value for all data bundles. For example, a data standard may apply to a single detector, in which case "detector number," although absent from all data bundles, can still be assumed to be present as a hidden potential dependency. Note in addition that, following earlier discussion, some constant-valued concept "A" may be linked to concept "B" in the data file purely by virtue of being in the same data bundle ("data block","data entry","file"); in such a case scientific knowledge may be required to identify the dependency. The simple question to ask is: "if 'A' had a different value, would (some values of) 'B' be different?"
Incorporating new datanames and domains into an ontological commons
New relationships arising from the above procedure may be as simple as an extra mapping between pre-existing types, for example, a soil temperature measured at a location where only air temperature had previously been measured (i.e., a new olog arrow from type "a location" to type "a temperature" labelled "has measured soil temperature"). A more complex situation arises when the new concepts involve an expansion or a restriction of pre-existing types, and it is in this case that our strictly functional relationships can be leveraged to our advantage. Consider the following example: Image ontology A contains the general type of "a frame" measured from "a detector" which consists of one or more "elements." Ontology B contains only the type "a frame measured from a single-element detector." In our olog view, we can construct the more restrictive types of Ontology B from the Ontology A types using "pullback" (also known as the "fiber product"). A set of objects created by a pullback is (isomorphic to) a set of pairs, with the elements of each pair coming from the two sets that have been "pulled back". The pairs appearing in the pullback set are restricted to those for which both elements map to the same element of a third set, so we are essentially filtering the set of all possible pairs. In mathematical terms, if f : A → C and g : B → C the pullback A × C B is {(a, b) | a ∈ A, b ∈ B, f(a) = g(b)}.
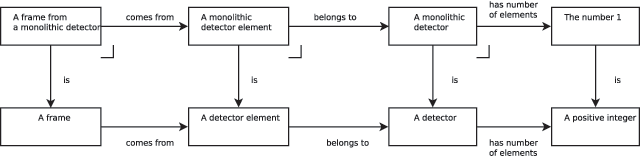
In our image ontology example, we can create the type "a monolithic detector" as the pullback of the type "detector" and single element type containing the integer "1", and the mappings "has number of elements" and "is." We can now continue to pullback (Figure 3) until we have arrived at the type "a frame from a single element detector." Usefully, mappings to the pulled-back types travel from the pulled-back type, which means that we have simultaneously specified the reverse operation: we can both compute the contents of the more restrictive datafile format, using the specification of pullback, and compute the contents of the more general datafile by simply following the mappings from the pulled-back types as usual. Of course, in a round-trip between the datafiles, any multi-element detector information is lost, as the more restrictive datafile bundle cannot contain this information.
|
An analogous operation to pullback ("pushforward") exists to fill in the bottom right-hand corner of our diagram given only the other three vertices; this is not described here but is well covered by Spivak & Kent[1] and Spivak.[16]
It is inevitable that an ontological commons created from multiple datafile specifications addressing related use cases will accumulate many overlapping definitions. It would be tedious and counter-productive if every subset of a type required complete definition of both that type and the possible datanames (e.g., "axis", "detector axis", "goniometer axis", "general axis" all might act as domains for the common mappings "axis vector", "axis offset", "axis position", "axis type"). Many of these multiple types are concisely specified using pullbacks, and as pullbacks explicitly define simple projections to the type of each member of the pair, the onward mappings defined for those types can be extended back to the pullback type as well; the convention can therefore be adopted in the ontology description that identical mapping names have identical meanings – provided that no name collisions occur for the two pulled-back categories. Appropriately equipped ontology description languages (such as OWL) can concisely model such pullbacks using the "class"-"subclass" relationship.
Application II: Machine translation between data formats
The early knowledge representation literature noted that an ontology can be used to mediate between differing data representations.[17] In terms of our definitions and ontological commons described above, data format translation involves transformation of values associated with datanames in dataname lists. Of course, a transfer of all data is only possible where the target dataname list contains the same, or more general, concepts as the source list.
The approach to scientific ontology described here deals with abstract types for values (e.g., "a real number"). Within a computer we do not have access to this abstract ontological space and so we must specify a value representation (e.g., "IEEE 754 floats represent real numbers," "all identifiers are character sequences") when using the ontology as the common ground for data file translation.
Given a canonical ontology created from source and target standards according to the procedure in Application I, the process of data file transformation is clear[g]:
- Identify a dataname list for each file type based on the datanames that appear, and whether or not multiple values are present.
- The intersection of the datanames appearing in 1 is a list of datanames whose values can be trivially obtained from the input file using an input-format-specific module. Note that to convert from multiple values for a given dataname to single values for that dataname, output a single data bundle for each of the multiple values. The reverse direction is trivially possible as a single value is a special case of multiple values.
- For each remaining item in the output list, use the aspects linking the types in the ontology to calculate the dataname values using as starting values the input file values obtained using the input-format-specific module.
- Use an output-format-specific software module to store the values calculated in 2 and 3.
If the values calculated at steps 2 and 3 are instead directly input into application software, that software becomes capable of reading any file format for which an input-format-specific module is provided.
The format-specific module
The format-specific module handling both the data item location and data item value conversions to common units and representation is a key component in the above translation procedure, essentially encapsulating the format adapter in machine-readable form. A suggested minimal interface (API) for a format-specific module follows, where we assume that one or more data bundles can be contained in a single file, and that no file is simultaneously open for reading and writing. Note that an object-oriented approach would hide the filehandle and datahandle parameters within an object, with these functions becoming methods of that object.
- get_by_name(datahandle,name,type,units)
- Return an array of values in optional
unitsin the agreed representation for the ontologicaltypeassociated with datanamenamein data bundledatahandle. Values returned for different names mapping from the same domain must occur at the same indices in the return array.
- set_by_name(datahandle,name,values,type,units)
- Set dataname
nameindatahandleto values of typetype. The same considerations as above apply to handling types and value correspondence.
- create_data_unit(filehandle,optional_name)
- Create and return a
datahandleto a new data bundle for filefilehandle.
- close_data_unit(datahandle)
- Finalize a data bundle. The data handle becomes invalid.
- open_file(filename,mode)
- Open a data file for reading or writing, returning a
filehandle.
- output_file(filehandle)
- Write out a file containing one or more data bundles.
The above functions include type and units parameters: type is included because the format adapter cannot, in general, deduce the appropriate ontological type given the format-specific value – this can be seen clearly for text-based formats such as CIF, where identical character sequences in a file could refer to a real number or to a simple text string (e.g., "1.1" might refer to Chapter 1, Section 1 or the real value 1.1). Likewise, providing units allows the implementation to transform the units used within the file to those prescribed by the ontology.
Machine-readable transformation instructions
Our simple ontology description leads to the simple, uniform nature of the values provided by the format-specific software modules, allowing any data transformations to be expressed using a uniform, file-format-agnostic language. Just as we are forced to choose a concrete representation for abstract ontological values, any mathematical algorithms found in the ontology must be transformed into an expression in a particular programming language using format- and programming language-specific interfaces before they can be used for data value transformation.
An extended example: Converting between NeXus and CIF raw image files
As a demonstration of many of the ideas presented here, code for transforming between files constructed according to the imgCIF[5] and NeXus/NXmx[4] standards has been developed (code supplied in supplementary material). Both of these standards relate to handling diffraction images produced by scanning single crystal samples in X-ray beams. CIF and NeXus use differing styles of format (custom text format and binary HDF5, respectively), and each standard targets radically different data bundles: a single imgCIF data bundle may contain multiple frames each with differing layouts; each frame may correspond to a collection of images from multiple detectors consisting of multiple modules. These frames can be obtained from multiple scans of multiple axes. By contrast, the scope of the NXmx definition is restricted to a single scan of a single axis, with each frame presented as a single image with unvarying structure.[h] Translating files in the presence of these dramatically varying scopes and formats is a useful test of the concepts presented here.
We follow the procedure outlined above to first determine a notional overall ontology for images that encompasses both NeXus and CIF.
(identifying types)
NeXus: NeXus includes the usual numerical types and arrays thereof, as well as character strings. In some cases character strings are drawn from a restricted set of possibilities. Each of these sets is a separate type.
CIF: The CIF2 standard includes integers, floating point numbers (with and without uncertainties), character strings and sets of character strings, homogeneous lists and arrays.
(identifying datanames)
NeXus: Broadly speaking, the NeXus specification[11] consists of "class" descriptions and "application definitions" built from those class descriptions. These classes have array-valued properties and attributes, and can be nested inside one another to form a hierarchy. A data bundle corresponds to a top-level root of the hierarchy, called an "entry" class. Multi-valued properties or attributes are constructed:
- using HDF5 arrays. These are denoted in the specifications using appended square brackets with a symbol inside, and datanames mapping from the same domain are indicated by use of the same symbol within the square brackets;
- by repeated use of a class;
- by encoding multiple values within a text string: for example, a single NeXus attribute lists multiple axis names in order of precedence.
(structural datanames) classes nested within another class have no significance beyond associating values (see Figure 4 and Discussion). Some items use array index as an implicit identifier for which a dataname should be created.
CIF: The CIF format directly associates either a single value or a column of values with a dataname appearing only once in the data bundle (called a "data block" in CIF). No other choices are available. Row position in a column is used only to indicate value correspondence.
A list of datanames used in the example is presented in Table 2. Intermediate datanames used during translation (including many of the aspects in Figure 3) are not shown. Translation of datanames useful to the human reader is also possible where simple equivalences exist (e.g., "sample identifier"), but have been omitted for brevity. Note that the datanames have been created purely for the purposes of this demonstration and have no other significance.
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The transformation code consists of three files written in Python: (i) a translation manager that is passed the required input/output formats and dataname list, calls the format adapters and ontology-based transformation routines, and splits the data bundles as necessary; and (ii) NeXus and CIF "format adapters" conforming to the API described above, which internally make use of third-party format-specific tools. Ontology-driven translation is accomplished via a machine-readable ontology written in a version of DDLm[18] enhanced to allow specification of pullback relationships. DDLm has been developed within the CIF community as a largely format-agnostic ontology language incorporating machine-readable dataname functional relationship specifications using dREL[19] The use of a different set of datanames for the API and the DDLm ontology is purely to demonstrate that datanames can be simple, readable text without the special symbols or formatting used in the pre-existing DDLm ontologies (known in the CIF framework as "dictionaries"). Indeed, the entire ontology could have instead been written in OWL, with embedded dREL methods, but the availability of DDLm dictionaries covering the subject area together with appropriate software made the current approach attractive. The translation manager internally translates between the canonical names and names used within the DDLm dictionaries.
To illustrate the roles played by the various components in the process of translation, the translation of a few representative datanames between CIF and NeXus is described in detail below. In each case, a list of datanames to be encapsulated in the output format is provided to the translation manager. Each dataname is either read directly from the input file, or calculated from relationships provided in the DDLm dictionary. The following descriptions elide some technicalities, which are covered in the documentation accompanying the supplementary material. In the following, a "category" is the DDLm term for the collection of datanames mapping from a single type and should not be confused with the "categories" of category theory or the closely similar "categories" of other ontological languages like OWL.
- incident wavelength (CIF to NeXus)
- The corresponding DDLm name in the DDLm ontology is consulted to determine the type ("Real") and units. The CIF format adapter is able to provide the associated values in the correct units using
CIF dataname _diffrn_radiation_wavelength.wavelength. The ontology specifies that dataname "incident wavelength" is a function of "wavelength id," which must therefore also be included in the output dataname list and is also provided directly by the CIF format adapter using identifier_diffrn_radiation_wavelength.wavelength _id. These two lists are passed to the NeXus format adapter which stores the wavelength values at hierarchical location NXinstrument:NXmonochromator:wavelength with canonical units and encodes "wavelength id" as the index of the value in this array.
- incident wavelength (NeXus to CIF)
- The wavelength is retrieved from the hierarchical location and the units converted to those requested, if necessary. The wavelength ids are generated as a sequence of integers corresponding to the position in the array of the corresponding wavelength. These two lists are passed through to the CIF format adapter. Note that a CIF-NeXus-CIF round trip is not guaranteed to preserve the textual values of wavelength id: this is acceptable, as the identifiers are opaque and arbitrary.
- simple scan data (CIF to NeXus)
- This canonical name has been assigned to the list of two-dimensional image frames collected from a single-element detector with uncoupled detector pixel axes and a corresponding DDLm name
_local_diffrn_scan_simple_frames.array_data.[i] The CIF format adapter cannot directly supply these image arrays, as the CIF file contains arrays that potentially correspond to individual elements from multi-element detectors and jointly-varying pixel axes. The translation manager therefore passes the CIF format adapter object to the ontology processing module with a request to derive "“simple scan data." The ontology contains the relationships of Figure 3 in machine-readable form, allowing the frame list to be automatically populated with only those frames that correspond to monolithic detector data, which is then passed to the NeXus format adapter. The NeXus format adapter stores the list of frame data at class location NXentry:NXinstrument:NXdetect-or:NXdata. "simple scan data" is a function of "simple scan data frame id," which is also derived by the ontology and, just as for wavelength, becomes encoded in the list position of the frames stored in the file.
- 2D data (NeXus to CIF)
- The NeXus image frames stored above are clearly a special case of the more general CIF frames. The ontology specifications are followed in the reverse direction to populate the canonical names recognised by the CIF format adapter. Just as for wavelength, the NeXus format adapter generates the frame identifiers from the frame positions, and these identifiers may not correspond to the original identifiers.
- detector axis vector mcstas (CIF to NeXus)
- This name refers to the direction of an axis associated with the detector, expressed in NeXus coordinates. The CIF format adapter cannot supply this, as CIF does not distinguish a separate detector axis type and uses a different coordinate system. The ontology defines the detector axis category as those items within the axis category with
_axis.equipmentequal to "detector" (i.e., a pullback), and the axis category (underaxis.local_vector_convention2) specifies how a NeXus axis vector can be obtained from a vector in the CIF coordinate system. These ontology methods are applied to obtain the requested vector values for all detector axes. These values are a function of "detector axis id." In this case, the axis ID is stored (as prescribed by NXmx) as a NeXus NXtransformations class group name, and the vector is stored as an attribute of this group. Note that the identifier text is explicit (and preserved) in this case as axis identifiers are used elsewhere in NeXus to describe each dimension of the data array.
- axis vector (NeXus to CIF)
- The ontology specifications are followed in reverse order to populate these values. In this case, the original axis identifiers are recovered.
The conceptual coverage of the ontology provided here is manifestly incomplete, as it is intended for demonstration purposes and as a check that each type of location and datavalue presentation in HDF5 is properly interpreted and constructed. While the code is freely licensed and thus may be used as a base for development, it should not be considered suitable for production purposes or bug-free. Note also that the datanames in the DDLm dictionary supplied are strictly for demonstration purposes and should not be used in any CIF data files unless they appear in official dictionaries. Further details of the transformation system are provided in the supplementary information.
Discussion
The "traditional" alternative to the ontology-based translation approach implemented above is to write dedicated software to perform direct translation between the two data standards. For example, Bernstein et al.[20] address the same imgCIF/NXmx translation problem by creating a "concordance," with equivalences and translations expressed in terms of the particular data structures of the respective file formats. This information is then consulted by programmers to write software specific to this particular pair of standards. Such an approach does not scale when the number of target formats increases, requiring N2 distinct translations for N formats, and judgements about equivalence and transformation algorithms are often (unlike the work of Bernstein et al.) hidden in computer code. The advantage of the approach described here is that ontological terms and relationships (that is, non-format-related information) are captured and accumulated in a universal system, which can be reused with other formats. For example, due to the broad scope of the imgCIF ontology and the significant expansion of that ontology entailed by the need to specify the more limited datanames appearing in the NXmx definition, virtually no further ontology development would be necessary to encompass the data appearing in many of the other, simpler, image formats. Therefore, it would be sufficient to provide a format adapter for a given image format (or class of formats) to allow immediate translation into any of the other formats already covered by a format adapter. This is simply a reiteration of the old argument that, given N standards, creating 2N translations to and from a single master standard is more efficient than N2 translations between each standard; the difference here is that the master standard is now a scientific ontology which cannot itself encapsulate data, but is accessible to the broader community and can thus be verified and developed.
The present work also underlines the point that complexity present in some data standards does not originate in the nature of the scientific information: the ontology demands only that a set of single or multi-valued datanames be present in a file with corresponding values correctly associated. Mechanisms designed to reduce file size are one source of complexity: for example, both HDF5 and imgCIF have image compression facilities.[j] Hierarchical formats save space by associating identifiers with node names and thereby reducing repetition, as demonstrated by comparing Table 3 (below) and Figure 4 (previous). This space-saving attribute of hierarchies becomes more effective as the number of datanames upon which a set of datanames depends increases, and is also attractive as an organizational tool: the reduced clutter in Figure 4 presumably allows the human brain to interpret the structure more rapidly.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Unnecessary complexity in the system arises from the indiscriminate use of complex structures available in a given format without an understanding of the simple essential ontological structure described here. Designers of data transfer standards are urged to consider that the fundamental task of any scientific data format is to associate multiple values to datanames; any complexity beyond that which is required for this purpose should be clearly justified in terms of the other goals of the format, such as efficiency, speed, or long-term archiving.
Conclusions
A simple formal approach using "ologs" has been applied to create a set of rules for constructing and curating a domain ontology. This framework was designed with accessibility to discipline experts as a key priority, while maintaining extensibility and modularity, and containing enough information to allow elaboration in formal ontological languages. As the tasks of expanding and merging these ontologies is entirely dependent on human intervention, this paper should not be counted as a significant contribution to the ontological mapping literature; from the perspective of knowledge representation researchers, the framework presented here does little more than describe some of the desirable consequences of adopting ologs as an ontological scheme. The primary significance of this work is, rather, in demonstrating to ontologically-naive scientific communities the value of a simple formal approach when developing and working with data standards: by following simple prescriptions, discipline experts can create ontologies that are guaranteed to be format agnostic, extensible, interoperable and accessible to other discipline experts; and the scientific content of legacy and current data formats can be seamlessly merged into these ontologies following a recipe. If the ontologies are then expressed in machine-readable form using one of a variety of ontological languages, such as OWL or DDLm, a modular, universal datafile input and translation system can be implemented without the need for an intermediate format to be defined.
Notes
- ↑ Note that "domain" and "codomain" are used throughout in the mathematical sense, as the set on which a function operates, and the set of resulting values, respectively.
- ↑ More strictly, datanames, the codomains of which this dataname maps from.
- ↑ A much shorter dataname such as iso.cross is, of course, also acceptable.
- ↑ OWL class-subclass relations could be used for this purpose.
- ↑ Among others, HDF5, the CIF format, relational databases and XML fulfill this requirement.
- ↑ Interestingly, heterogeneous lists of values are unlikely to be needed in data files, as any processing of such a list will by definition require separate treatment for each element, and consequently a distinct and definable meaning is available for the individual components which can thus be included in their own right within the ontology. While the ontology itself will still contain such heterogeneous tuples (e.g., for a dataname that is a function of multiple other datanames with differing types), the data file itself does not need to contain this tuple so long as the tuple can be constructed from its individual components on input, if necessary, by the programmer.
- ↑ This approach to translation assumes that the dataname lists are drawn from a common ontology, which is predicated on human intervention to combine the differing ontologies. Spivak[13] treats in detail a situation in which ontologies remain separate, but a morphism between the ontologies is stipulated (a "functor"). While such a morphism can generally be computer-derived in the context of database transformations, in the present discussion human intervention is required and therefore creation of a single common ontology is the simpler route.
- ↑ Conformant NXmx files can be produced with a somewhat broader scope than that described here. A restricted scope has been chosen for the purposes of demonstration. Interested readers are directed to the NeXus website (www.nexusformat.org) for more information.
- ↑ In accordance with CIF convention, all unofficial datanames contain the string "local."
- ↑ Images as two-dimensional arrays of numbers are themselves an efficient representation of the basic olog function taking a pair of integers (the horizontal and vertical pixel coordinates) to a number (the intensity), where the efficiency arises by having the pixel coordinates encoded in the position of the number in the overall array.
Supplementary files
The supplementary files for this article can be found as follows:
Supplementary File 1: Appendix: http://dx.doi.org/10.5334/dsj-2016-012.s1
Supplementary File 2: Software distribution: http://doi.org/10.5281/zenodo.154459
Competing interests
The author declares that they have no competing interests.
References
- ↑ 1.0 1.1 1.2 Spivak, D.I.; Kent, R.E. (2012). "Ologs: A categorical framework for knowledge representation". PLoS One 7 (1): e24274. doi:10.1371/journal.pone.0024274. PMC PMC3269434. PMID 22303434. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3269434.
- ↑ Boulton, G. (2012). "Open your minds and share your results". Nature 486 (7404): 441. doi:10.1038/486441a. PMID 22739274.
- ↑ Kroon-Batenburg, L.M.; Helliwell, J.R. (2014). "Experiences with making diffraction image data available: What metadata do we need to archive?". Acta Crystallographica Section D Biological Crystallography 70 (Pt. 10): 2502-9. doi:10.1107/S1399004713029817. PMC PMC4187998. PMID 25286836. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4187998.
- ↑ 4.0 4.1 "NXmx – Nexus: Manual 3.1 documentation". NeXusformat.org. NIAC. 2015. http://download.nexusformat.org/doc/html/classes/applications/NXmx.html.
- ↑ 5.0 5.1 Bernstein, H.J. (2006). "Classification and use of image data". International Tables for Crystallography G (3.7): 199–205. doi:10.1107/97809553602060000739.
- ↑ "Detectors & Formats recognized by the HKL/HKL-2000/HKL-3000 Software". HKL Research, Inc. 2016. http://www.hkl-xray.com/detectors-formats-recognized-hklhkl-2000hkl-3000-software.
- ↑ Otero-Cerdeira, L.; Rodríguez-Martínez, F.J.; Gómez-Rodríguez, A. (2015). "Ontology matching: A literature review". Expert Systems with Applications 42 (2): 949–971. doi:10.1016/j.eswa.2014.08.032.
- ↑ Shvaiko, P.; Euzenat, J. (2013). "Ontology matching: State of the art and future challenges". IEEE Transactions on Knowledge and Data Engineering 25 (1): 158–176. doi:10.1109/TKDE.2011.253.
- ↑ Hitzler, P.; Krötzsch, M.; Parsia, B. et al. (11 December 2012). "OWL 2 Web Ontology Language Primer (Second Edition)". W3C Recommendations. W3C. https://www.w3.org/TR/2012/REC-owl2-primer-20121211/.
- ↑ Hall, S.R.; McMahon, B., ed. (2005). International Tables for Crystallography Volume G: Definition and exchange of crystallographic data. Springer Netherlands. doi:10.1107/97809553602060000107. ISBN 9781402042904.
- ↑ 11.0 11.1 Könnecke, M.; Akeroyd, F.A.; Bernstein, H.J. et al. (2015). "The NeXus data format". Journal of Applied Crystallography 48 (Pt. 1): 301–305. doi:10.1107/S1600576714027575. PMC PMC4453170. PMID 26089752. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4453170.
- ↑ Henley, S. (2006). "The problem of missing data in geoscience databases". Computers & Geosciences 32 (9): 1368–1377. doi:10.1016/j.cageo.2005.12.008.
- ↑ 13.0 13.1 Spivak, D.I. (2012). "Functorial data migration". Information and Computation 217: 31–51. doi:10.1016/j.ic.2012.05.001.
- ↑ Strickland, P.R.; Hoyland, M.A.; McMahon, B. (2005). "5.7 Small-molecule crystal structure publication using CIF". In Hall, S.R.; McMahon, B.. International Tables for Crystallography Volume G: Definition and exchange of crystallographic data. Springer Netherlands. pp. 557–568. doi:10.1107/97809553602060000107. ISBN 9781402042904.
- ↑ "Hierarchical Data Format, Version 5". The HDF Group. 2016. https://support.hdfgroup.org/HDF5/.
- ↑ Spivak, D.I. (2014). Category Theory for the Sciences. MIT Press. pp. 496. ISBN 9780262028134. http://category-theory.mitpress.mit.edu/index.html.
- ↑ Gruber, T.R. (1993). "A translation approach to portable ontology specifications". Knowledge Acquisition 5 (2): 199–220. doi:10.1006/knac.1993.1008.
- ↑ Spadaccini, N.; Hall, S.R. (2012). "DDLm: A new dictionary definition language". Journal of Chemical Information and Modeling 52 (8): 1907-16. doi:10.1021/ci300075z. PMID 22725613.
- ↑ Spadaccini, N.; Castleden, I.R.; du Boulay, D.; Hall, S.R. (2012). "dREL: A relational expression language for dictionary methods". Journal of Chemical Information and Modeling 52 (8): 1917-25. doi:10.1021/ci300076w. PMID 22725641.
- ↑ Bernstein, H.J.; Sloan, J.M.; Winter, G. et al. (10 May 2014). "Coping with BIG DATA Image Formats: Integration of CBF, NeXus and HDF5, A Progress Report" (PDF). American Crystallographic Association Meeting, 24–28 May 2014. American Crystallographic Association. https://sites.google.com/site/nexuscbf/home/presentation-foils/Integration_Poster_10May14_w_overlays.pdf.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. The original article lists references alphabetically, but this version — by design — lists them in order of appearance. Footnotes have been changed from numbers to letters as citations are currently using numbers. Many of the internal references to section numbers were removed as section numbers were not retained, largely due to LIMSwiki title formatting rules.