Journal:SODAR: Managing multiomics study data and metadata
| Full article title | SODAR: Managing multiomics study data and metadata |
|---|---|
| Journal | GigaScience |
| Author(s) | Nieminen, Mikko; Stolpe, Oliver; Kuhring, Mathias; Weiner III, January; Pett, Patrick; Beule, Dieter; Holtgrewe, Manual |
| Author affiliation(s) | Berlin Institute of Health at Charité–Universitätsmedizin Berlin |
| Primary contact | Email: mikko dot nieminen at bih dash charite dot de |
| Year published | 2023 |
| Volume and issue | 12 |
| Article # | giad052 |
| DOI | 10.1093/gigascience/giad052 |
| ISSN | 2047-217X |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://academic.oup.com/gigascience/article/doi/10.1093/gigascience/giad052/7232111 |
| Download | https://academic.oup.com/gigascience/article-pdf/doi/10.1093/gigascience/giad052/50974561/giad052.pdf (PDF) |
Abstract
Scientists employing omics in life science studies face challenges such as the modeling of multiassay studies, recording of all relevant parameters, and managing many samples with their metadata. They must manage many large files that are the results of the assays or subsequent computation. Users with diverse backgrounds, ranging from computational scientists to wet-lab scientists, have dissimilar needs when it comes to data access, with programmatic interfaces being favored by the former and graphical ones by the latter.
We introduce SODAR, the system for omics data access and retrieval. SODAR is a software package that addresses these challenges by providing a web-based graphical user interface (GUI) for managing multiassay studies and describing them using the ISA (Investigation, Study, Assay) data model and the ISA-Tab file format. Data storage is handled using the iRODS data management system, which handles large quantities of files and substantial amounts of data. SODAR also offers programmable application programming interfaces (APIs) and command-line access for metadata and file storage.
SODAR supports complex omics integration studies and can be easily installed. The software is written in Python 3 and freely available at https://github.com/bihealth/sodar-server under the MIT license.
Keywords: omics, research, research data, data management
Introduction
Modern studies in life sciences rely on “omics” assays, which encompass branches of science such as genomics, proteomics, and metabolomics. One or multiple assays can be run within a single study, potentially including assays for multiple omics studies of several types.
The following key steps are required for executing these complex omics studies: (i) planning, which results in study metadata; (ii) collection of mass data; and (iii) data analysis, including the integration of multiple assays. The aim of SODAR is to ensure support for scientists within all the steps.
Challenges
Each step presents its own set of challenges. During planning, it is important to enable recording crucial factors and covariates. The flow of materials and samples through processes must also be specified in sufficient detail. Further challenges arise from, for example, assays using complex multiplexing, such as the need for reference samples; requirements for using controlled vocabularies or ontologies; and possible change of assays over time.
In the data collection step, scientists must record the used machines, kits, and versions of both hardware and software used. Omics studies also create large volumes of data, ranging from a few gigabytes for mass spectrometry (MS) to terabytes for imaging such as microscopy. These data may be spread among many files, further complicating the needs for managing mass data storage. Instead of a rigid process, data collection should also be adjustable to changes and developments in data generation over time.
Data analysis is often split into multiple phases, with primary analysis of each assay followed by steps for integration of results. Specific results need to be fed back to metadata management, annotation, quality control, or storing resulting markers. Access to metadata with recorded factors and confounders is necessary in each step, while access to primary raw data becomes less important after the primary analysis. Certain analysis results are written back into the mass data storage. This includes binary alignment map (BAM) files and variant call format (VCF) files.
There are also overarching challenges for the steps in study execution. All data should be recorded in structured format. Automation should be applied where possible, and on-premises installation might be preferable or even required when data privacy–relevant data are generated such as with DNA sequencing.
Data management approaches
In this and the following section, we will discuss the topic of |data management and software. The terms “data” and “document” will be used interchangeably in this section. The steps described in the prior subsection can be interpreted as processes taking documents and materials as input, as well as generating more documents and materials as the result. For example, data collection takes the plan document and samples and generates assay result files (documents). Scientists thus need computational tools for supporting them in managing their scientific and research data.
Historically, such documents are maintained on paper in laboratory notebooks, or documentation created by quality management systems (QMSs). For the most direct and unstructured approaches in maintaining digital data, this corresponds to word processing, spreadsheet, and image files on local or network drives. More structured approaches are desirable for taking advantage of digital documents, preventing research data loss[1] or fostering reuse.[2]
While data management in science is a broad topic, the library and information science community is frequently approaching it using a top-down approach. Frequently, in this context, the term “research data management” (RDM) is used. Here, the needs of whole organizations and their parts for managing their research data, as well as the necessary steps to establish whole RDM systems, are considered first (cf. Donner[3]). This correlates with the role of libraries in certain academic organizations for organizing data that were collected in research.
A second approach, which can be described as “bottom-up,” originates from different “working scientist” communities. The communities commonly refer to the topic as “scientific data management” (SDM) and solve their problems at hand, often starting with specific small-scale solutions, which are then upscaled if the need arises. While considering their organizational embedding, they focus on solving specific data management challenges for themselves and their peers. We found ourselves in this situation and will thus focus on this perspective.
Data management software packages
SDM needs come in different forms and shapes. We could find no general treatment of the subject of data management in the literature. Machina and Wild[4] provide a collection of four tool categories: laboratory information management systems (LIMSs), electronic laboratory notebooks (ELNs), scientific data management systems (SDMSs), and chromatography data systems (CDSs) that we generalize as instrument-specific data systems (IDS). In this section, we provide our take on explaining what these systems comprise. We also note—as Machina and Wild[4] did—that categorization of such software solutions is not clear-cut, and features may be overlapping. We expand this list by two more system types: data repository systems (DRSs) and database/data warehouse management frameworks (DMFs).
The four tools described by Machina and Wild[4] are as follows:
- A LIMS focuses on storing information around laboratory workflows. This includes tracking of consumables, samples, instruments, and tests. A LIMS deals with daily tasks of laboratories such as billing and instrument calibration. It is often specific to certain domain areas such as sequencing facilities.
- The ELN focuses on allowing humans to record their laboratory work. It replaces paper notebooks and captures experiments and their results, mostly in free-form text, pictures, tables, and so on. The ELN plays a key role in fulfilling regulatory requirements.
- The CDS/IDS provides data capturing, storage, and analysis functionality in instrument-specific domains. Two examples are the CASAVA pipeline and the BaseSpace cloud-based service, both from Illumina. The former is provided without extra cost with the instrument along with its source code, while the latter is purchasable and closed source. Such software often ships with the instruments themselves.
- The SDMS provides scientific content management functionality for scientific data and documentation. It allows for the management of metadata and potentially mass data. The SDMS's core functionality generally doesn't include data analysis, user-centric data collection, or laboratory workflow tracking. Such features may be potentially supported by plugins or extensions. Many such systems offer integration with surrounding systems (e.g., via application programming interfaces [APIs]).
We augment this list by two system types:
- The DRS provides shared access to data with appropriate documentation and metadata. Examples are FAIRdom Seek[5], Dataverse[6], and Yoda.[7] There are also specialized DRSs focusing on particular use cases, such as dbGAP[8], MetaboLights[9], and Gene Expression Omnibus[10], that allow for managing public or controlled public access to large research data collections.
- The DMF allows for the rapid development of database and data warehouse applications. It often provides preexisting components to build on ready-made functionality and extension by implementing custom components. This enables the creation of domain-specific databases and structured data capturing. Examples include Molgenis[11] and Zendro.[12]
Other types of systems also exist, and not every system falls into just one category. A complete review of such systems is beyond the scope of this article. This section identifies focus areas of systems involved in some form of scientific data management. SODAR falls into the category of SDMS.
Data management technologies
For planning and documenting experiments and their structure, experiment-oriented metadata storage formats with predefined syntax and semantics exist. A popular standard is the ISA (Investigation, Study, Assay) model[13], which allows describing studies with multiple samples and assays. The ISA model defines the ISA-Tab tabular file format, which allows users to model each processing step with each intermediate result and annotate each of these with arbitrary metadata. An example of an alternative to ISA-Tab is Portable Encapsulated Projects (PEPs).[14] There are also more specialized standards such as Brain Imaging Data Structure (BIDS) for brain imaging data[15], as well as other approaches such as Clinical Data Interchange Standards Consortium (CDISC) standards[16] and the Hierarchical Data Format (HDF5).[17] Use of generic file formats such as HDF5, TSV, XML, and JSON is also common.
For storing large volumes of omics data, it is possible to simply use file systems or object storage systems. More advanced solutions such as Shock[18] or dCache[19] allow for storing metadata and distributing data over multiple servers. iRODS (Integrated Rule-Oriented Data System)[20][21] adds further features, such as running rules and programs within the data system and enabling integration with arbitrary authentication methods.
For publication, raw and processed data and metadata are deposited in scientific catalogs, study databases, and registries. Examples include the BioSamples database for metadata[22] and Sequence Read Archive (SRA) for raw sequencing data.[23]
Our work
In our work, we focus on managing many omics projects of varying data size and various use cases, including cancer and functional genomics studies. We also need to support multiple technologies such as whole genome sequencing, single-cell sequencing, proteomics, and MS. Deposition to public repositories was not a necessity in our context. However, SODAR is an ISA-compliant system. Should the data owner require it, it is easily feasible to create appropriate exports to public data repositories using the APIs provided by SODAR. Open-source software is a requirement to avoid vendor lock-in and allow for flexibility in different use cases. A suitable end-to-end solution was not available when we started our work in 2016. Therefore, we set out to implement an integrated system for managing omics-specific data and metadata.
In this article, we introduce SODAR (System for Omics Data Access and Retrieval). SODAR combines the modeling of studies and assays using the ISA-Tab format with handling of mass data storage using iRODS. More example projects are available in the SODAR online demo server.[24]
Results
We present the results by first giving an overview of the developed SODAR system. Next, we compare it to a selection of existing tools and their relevant features. We then describe processes we have established around SODAR. Finally, internal usage statistics are detailed along with discussion on the limitations of SODAR.
Resulting system overview
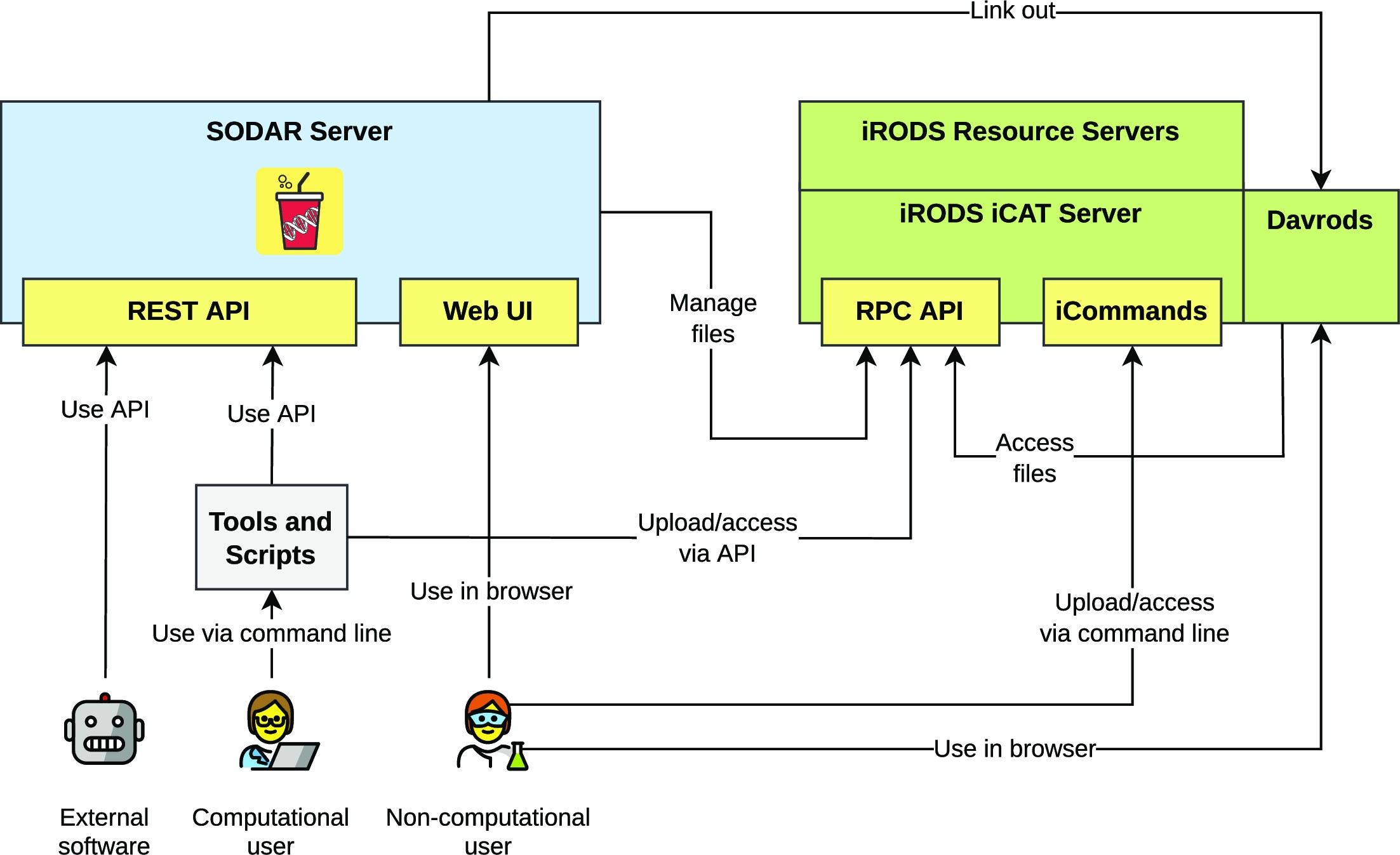
Figure 1 presents the components of the SODAR system. The SODAR server is built on the Django web framework. It contains the main system logic and provides both a graphical user interface (GUI) and APIs for managing projects, studies, and data.
|
Project and study metadata are stored in a PostgreSQL database. The study metadata are stored as ISA-Tab–compatible sample sheets, with each project containing a single ISA-Tab investigation. Each investigation can hold multiple studies; likewise, each study can contain multiple assays.
Mass data storage is implemented using iRODS and accessed via iRODS command-line tools or access to the WebDAV protocol, which is provided by using the Davrods software. The SODAR server manages creation of expected iRODS collections (i.e., directories), governs file access, and enforces rules for file uploads and consistency. Investigations, studies, and assays correspond to collections in the iRODS file hierarchy. Within assays, collection structure can be split by, for example, samples or libraries, depending on the type of assay.
Uploading files for studies is handled using “landing zones,” which are user-specific collections with read and write access. The SODAR server handles validation and transfer of files from the landing zones into the project-specific read-only sample repository, which is split into assay-specific iRODS collections.
Planning and tracking the study design and experiments is done using the ISA-Tab–compatible sample sheets. Here, the “assay” in the ISA model corresponds to an “experiment” in our work. SODAR provides multiple ways to create and edit both the metadata model and the contained metadata itself, including user-friendly GUI-based creation of sample sheets from ISA-Tab templates. The templates aid in maintaining consistent metadata structures between studies. Once created, the SODAR server provides a GUI for filling up metadata and configuring expected values, including support for controlled vocabularies and ontologies. Furthermore, SODAR also allows uploading and updating sample sheets using its API. Uploading any valid ISA-Tab file and replacing existing sheets via upload is also supported, enabling the creation of sample sheets using other software such as ISA-tools.[13] The API allows to automate metadata and file management activities using scripts.
Data management software features and selection
This section first describes features of DMS packages that are subsequently used for comparing SODAR to other software types and packages. We then describe the selection process for software comparison.
The following is a list of features that allows us to see the unique strengths and properties of SODAR within the scope of an SDMS and describe SODAR's difference from other data management systems. When a feature is important in multiple categories, it is only shown once. Feature categories 1–4 are focused on SDMS, and category 5 contains features also important for other data management systems.
- 1. Features addressing overarching challenges
- a. Structure into projects and folders
- b. Access control
- b. Automation possible via API
- 2. Use of open formats and standards
- a. Features addressing planning challenges
- b. Structured recording of assays and experiments
- c. Flexibility in definition of studies and experiments
- d. Annotation with controlled vocabulary
- e. Annotation with ontologies
- 3. Features addressing data collection challenges
- a. Storage of files possible
- b. Support for many files
- c. Support for large file sizes
- 4. Features addressing data analysis challenges
- a. API for reading and updating experiment metadata
- b. API for reading and updating mass data
- 5. Features commonly found in specific systems
- a. ELN
- i. Flexible data entry in free text/tables/pictures
- b. DRS
- i. Host public data repositories
- c. DMF
- i. Easy creation of new data tables
- ii. User-centric data entry
- iii. Multiple predefined components (e.g., for data visualization and analysis)
- a. ELN
With the aim of showing the unique strengths of software categories and packages, we attempted to select popular software packages in each category. We limited the selection to open-source software. We searched for the different software types via a publication on Google Scholar or the project search on GitHub. We made no attempt to define “the most popular” or “the best” software packages. We excluded LIMS and IDS as such software is focused on the wet-lab process. The following software was selected:
- 1. SDMS
- 2. ELN
- 3. DRS
- 4. DMF
Data management software comparison
The table included in Additional File 1 shows the comparison of the categorized software in the categories as described in the previous subsection.
Since the software packages operate in a similar space, there is a certain overlap in features, even across system types. Most software packages provide the features for addressing the overarching challenges. All “planning” features are included in SODAR and FAIRDom Seek in the SDMS category, while qPortal and OpenBIS remain limited. ELabFTW provides limited functionality for structured recording and does not support controlled vocabularies and ontologies, while DRS systems do not address planning challenges by their design. As expected, such features can be implemented by the DMF packages, but they do not provide the functionality on their own. The “data collection” and “data analysis” features are only comprehensively addressed by SODAR and FAIRDom Seek in the SDMS category, with FAIRDom Seek being limited in storing many and/or large files. ELN software is limited in this capability, while DRS packages provide good support for such features, and the DMF software packages allow for implementing support to varying levels.
As for the specialized features, some functionalities of “foreign” categories are implemented. For example, SODAR has support for user-centric data entry, and FAIRDom Seek allows for hosting public data repositories by design. However, each software package shows its strengths by providing the features for the tasks that it was originally designed for. We note that certain packages cover their category more focused or comprehensively than others. For example, in the DMF category, Molgenis has an ecosystem of many predefined components, while Zendro focuses on allowing for the easy creation of tables and user-centric data entry masks.
Roles and interaction with SODAR
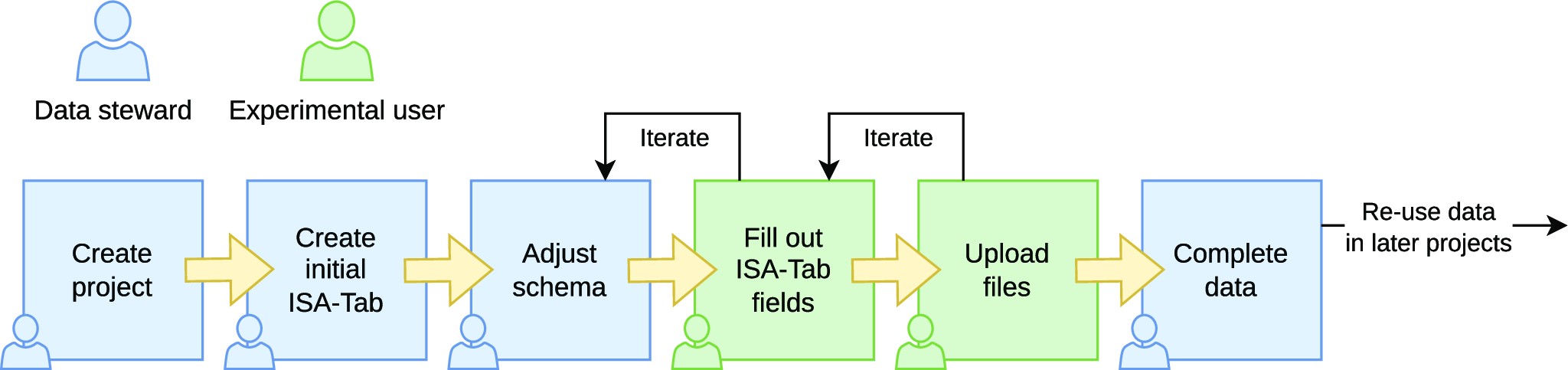
The general workflow in using SODAR for managing data and metadata is shown in Fig. 2. We distinguish between the roles “data steward” and “experimentalist.” It is possible for one person to act in both roles.
|
Data stewards are responsible for creating the overall structure of the experiment data. They are expected to be experienced with using ISA-Tab files. For example, in our use case, data stewards are bioinformaticians working in the core unit. They are responsible for planning the experiments and modeling them in the ISA-Tab format as sample sheets describing the overall experimental design. Data stewards also maintain a library of sample sheet templates for common use cases. With experienced experimentalists, the steward might just create the general structure of the experiment. In some cases, the steward may also pre-create the sample sheet with an initial structure of all planned samples and processes and IDs together with experimentalists.
Experimentalists are primarily responsible for entering the actual data into the system. They are users more concerned with completing the metadata in the sample sheet than in creating its structure. When the full sample sheet is created together with data stewards, experimentalists may only verify the structure against the information of their experiments and fill in some measurements in sample sheet cells (e.g., concentration measurements). More experienced experimentalists will also create new rows in the ISA-Tab tables for samples, related materials, and processes.
General SODAR process
Here we describe the SODAR-backed process of managing experiment data we are using in our work. This demonstrates how SODAR helps tackle challenges in complex omics study management.
Planning and sample sheet creation
Planning begins with data steward and experimentalists meeting and discussing the study, including, for example, its factors, sample size, replicas, and confounders. Stewards create sample sheets from templates and modify columns depending on the discussions and the study's requirements. Working together, stewards and experimentalists also decide on ontologies and controlled vocabularies to use, data ranges, and so on.
The template will be bootstrapped with example samples, or all samples, depending on the study. During this step, the experimentalist receives training in using the SODAR sample sheet editor for filling in cells where necessary. Filling cells can involve, for example, adding measurements, cancer staging, definition and refinement of phenotypes, and adjustment of relationship information.
Automated extraction of measurements from instruments or LIMS and ingesting it using the SODAR API is also possible. For example, an integration with a LIMS system could automatically create samples as they are processed in the wet lab, while measurements could be written to SODAR from the LIMS or from an integration of an ELN system. We are currently working toward this when cooperating with other units.
Data acquisition and sample sheet update
Experimentalists run their experiments and use SODAR for editing the sample sheets. This includes adding new samples, marking dropouts, or removing them, as well as adjusting ontologies and terms as needed. SODAR sample sheets are useful as a central storage of metadata, removing the need to, for example, share spreadsheets via email. Differences between sample sheet versions can also be browsed in the SODAR GUI to track changes in the metadata.
In this step, actual data files are uploaded by experimentalists to the project sample repository through landing zones. The iRODS collection structure for each study is maintained by SODAR and based on the study type and names of samples or associated libraries. In most cases, files related to a certain sample and its processing in an assay can be found in the collection named after the related library.
Data analysis
For data analysis, bioinformaticians access metadata in the sample sheets as well as raw data in iRODS, the latter being linked to the former in the SODAR GUI for ease of access. Depending on the phase of study, this may involve, for example, primary analysis, secondary analysis, and required data integration. Resulting files are uploaded back into iRODS via SODAR for safekeeping and sharing between researchers. Also, uploaded are files needed for integrating with third-party systems, such as UCSC Genome Browser[28] tracks and files for data exploration tools such as SCelVis.[29]
During the analysis, up-to-date experiment structure is maintained in SODAR. It represents a centralized storage and sole source of truth for the internal structure, encompassing factor values, ontologies, and controlled vocabularies. Similarly, it represents an external structure, with samples and materials linked to corresponding iRODS collections.
SODAR also provides integrations to specific third-party software to aid analysis. For germline and cancer DNA sequencing experiments, SODAR supports the IGV Genome Browser[30], by generating session files pointing at relevant variant and read alignment files with a single click.
Long-term data storage and data access
After transferring files from landing zones into the project's sample repository, the data are in general assumed to be permanent and not modifiable or rewritable, with users only having the possibility of request file deletion from project maintainer in case of, for example, mistakes in uploading. Hence, once the project finishes, the data are considered good for long-term archival use. SODAR supports setting projects into a read-only “archived” state and provides an API for implementing custom policies for handling archived data. For example, such a policy might consist of adding a cold storage resource such as tape onto which the data could be moved.
In exporting data to public databases, creating a generic exporter cannot be considered feasible due to the metadata model flexibility in SODAR. However, there are export possibilities depending on the type of study. For example, if the project is set up with Gene Expression Omnibus (GEO)[10] compatible metadata, exporting to the GEO database may be trivial depending on the target system APIs. In the future, we intend to create export functionality from SODAR to the emerging German National Research Data Infrastructure (NFDI), the associated German Human Genome-phenome Archive (GHGA)[31], and corresponding metadata models. These will be based on the federated European Genome-phenome Archive (EGA)[32] and should provide a good starting point for many other exporters. NFDI will be our long-term and controlled public access backend, while other users and instances might have other backends.
Internal usage statistics
We have been using SODAR in our group's projects for the past 4 years. Table 1 summarizes data statistics and metadata stored in our internal instance and the diversity of projects. We have thus tested SODAR extensively in a real-world setting and use it daily as our main storage for all our project data and metadata.
| ||||||||||||||
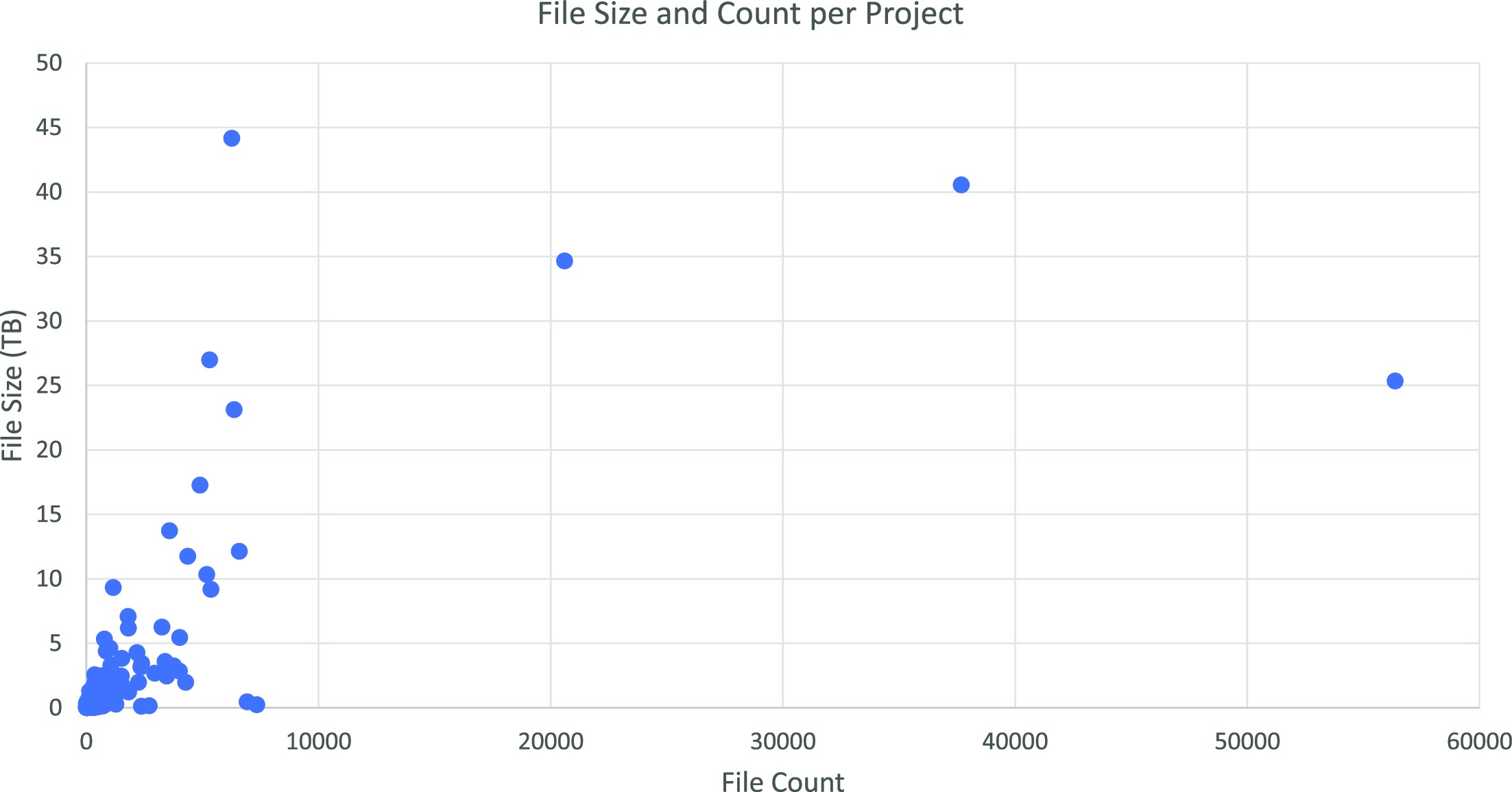
Figure 3 displays file size and count for each project on our system in March 2022. The diagram shows the varying scale of the projects within our group. A limited number of projects from a 20- to 45-terabyte range can be seen, while most are smaller.
|
Limitations
Currently, SODAR offers no automated data export to, for example, the GEO database. This may be added in the future as discussed in the “Long-term data storage and data access” section. Similarly, SODAR does not support access in a “data commons” manner. It is possible to set specific projects for public read access, but by default, SODAR enforces strict access control to data.
We also do not have a definitive solution for training people in ISA-Tab. SODAR features a set of templates for predefined study types (e.g., germline and cancer studies), but there is no definite solution for trivially setting up any type of study as ISA-Tab.
Methods
SODAR (RRID:SCR_022175) is implemented in Python 3 using the Django (RRID:SCR_012855]) web framework and Django REST Framework. Reusable components have been extracted into the library SODAR Core (RRID:SCR_023708). ISA-Tab format manipulation has been implemented using AltamISA (RRID:SCR_023709).[33]
Project organization, authorization structure, and LDAP integration
SODAR uses the concept of “projects” for organizing all data. Projects have a unique identifier and some basic metadata, such as title and description. Projects are organized in a tree structure using the concept of “categories” that can contain projects or other categories. Each project has a single owner, who can assign themselves a delegate for managing the project. Further users can be granted access to the project either in a read-write (contributor) or a read-only fashion (guest) using role-based access control (RBAC).[34]
SODAR can be configured to be run standalone or integrated with LDAP servers, including Microsoft ActiveDirectory, for providing authentication information. Here, authentication refers to checking the identity of a user based on their username and password.
iRODS integration
SODAR automatically manages user access to projects in iRODS. This is done by creating an iRODS directory and user group for each project. The group is given access to the directory, and group membership is synchronized between the SODAR database and iRODS.
SODAR creates an iRODS collection for each study and assay from the ISA model of the project. Files can be uploaded by users through landing zones, either for each sample or for the whole study or assay. It is thus possible to add data for an arbitrary number of assays for each sample and original donor or specimen.
The files can be accessed either directly through iRODS or using the WebDAV protocol through the Davrods[35] software. The latter allows users to access the storage as a network drive on their desktop computers. Since WebDAV is HTTP-based, users can also make data available to genome browsers such as the Integrative Genomics Viewer (RRID:SCR_011793) or UCSC Genome Browser (RRID:SCR_005780). Moreover, it is easy to access data through an organization's security system and proxies without the intervention of IT departments.
Optionally, SODAR allows the management of iRODS “tickets,” which allow for access based on randomly generated tokens instead of user login. This way, users can upload genome browser tracks to SODAR and iRODS and create public URL strings to access them and share them with users that do not have access to the full project or do not even have an account in SODAR.
Sample sheet editor, import, export
Sample sheets can be included into SODAR projects by either importing existing ISA-Tab files or template-based creation. When importing, the user can upload a Zip archive or a set of individual ISA-Tab files. For creating sample sheets from templates, the user needs to fill in certain details in the SODAR GUI. SODAR contains multiple built-in templates for generic RNA sequencing, germline DNA sequencing, and MS–based metabolomics, for example. After import or creation, the sample sheets are stored in an object-based format in the SODAR database for easy search and modification. In the GUI, they are presented to the user as spreadsheet-style study and assay tables.
The user can edit sample sheets in the SODAR GUI (see Additional File 2). Cells in the study and assay tables can be edited like in a spreadsheet application. For each column, the project owner or delegate can define the accepted format, value choices, value ranges, regular expressions for accepted values, and other settings depending on the column type. This ensures the validity of data and their compatibility with the study's requirements and conventions.
SODAR supports ontology term lookup for cell editing. Commonly used ontologies such as Human Phenotype Ontology (HPO) (RRID:SCR_006016)[36], Online Mendelian Inheritance in Man (OMIM) (RRID:SCR_006437)[37], and NCBI Taxonomy Database Ontology (NCBITaxon) (RRID:SCR_000479)[38] can be uploaded into SODAR for local querying as OBO or OWL files, without the need to rely on third-party APIs. Manual entering of ontology terms is also allowed. It is possible to include multiple ontology terms in a single cell, and one or several ontologies can be used in a single column.
When editing sample sheets, old sheet versions are stored as backup. These versions can be compared and restored in case of mistakes, as well as exported from the system. SODAR allows for sample sheet export in the full ISA-Tab TSV format or simplified Excel tables. Replacing existing sheets with versions modified outside of SODAR is also supported.
Integrating SODAR Core–based sites
Several subcomponents of the SODAR server such as project and user management have proven to be useful in other contexts. We have extracted them into the SODAR Core software package[39], which forms the foundation of other projects such as VarFish (RRID:SCR_023710)[40] and Kiosc (RRID:SCR_023711).[41] Using a common library for projects and access management has several advantages and enables the integration of VarFish and Kiosc with SODAR.
SODAR can be configured to work as a “source” site. Applications based on SODAR Core can then be configured as “target” sites of the source site. Projects and access to users will then be synchronized to target sites. This allows us to manage sample and experiment definitions in SODAR and upload corresponding variant data to VarFish. VarFish can then use the REST APIs defined by SODAR for synchronizing sample metadata, such as phenotype terms, directly from SODAR. Similarly, users can upload mass data files into the iRODS data repository and create access tokens to them in SODAR. These tokens can be used to provide data visualization applications in Kiosc with data access via HTTP and iRODS protocols or external applications such as UCSC Genome Browser.
SODAR administration
We provide a straightforward way to install SODAR and related components (SODAR, iRODS, Davrods, and supporting database servers) and maintain such an installation based on Docker containers and Docker compose. Detailed installation instructions can be found in the “sodar-server” source code repository.[24]
The entire system can be set up using an external LDAP or ActiveDirectory server for users and credentials or as an alternative in a standalone fashion where SODAR hosts this information. Existing iRODS installations can also be used with SODAR. For administrators, SODAR features dashboards that provide statistics regarding projects and usage of storage resources.
Availability of supporting source code and requirements
Project name: sodar-server
Project homepage: https://github.com/bihealth/sodar-server
Operating system: Linux/Unix
Programming language: Python
License: MIT
RRID: SCR_022175
Biotools: biotools:sodar
Additional Files
The following additional files can be found in giad052_supplemental_files.zip:
- Additional File 1. Data management software comparison table. Comparison of features between SODAR and related data management software
- Additional File 2. SODAR sample sheet editor. Figure consisting of screenshots of the SODAR sample sheet editor with its major features annotated
Abbreviations, acronyms, and initialisms
- API: application programmable interface
- BAM: binary alignment map
- BIDS: Brain Imaging Data Structure
- CDISC: Clinical Data Interchange Standards Consortium
- DMF: data management framework
- DRS: data repository system
- EGA: European Genome-phenome Archive
- ELN: electronic laboratory notebook
- GEO: Gene Expression Omnibus
- GHGA: German Human Genome-phenome Archive
- GUI: graphical user interface
- HDF5: Hierarchical Data Format v5
- HPO: human phenotype ontology
- IDS: instrument-specific data system
- iRODS: Integrated Rules-Oriented Data System
- ISA: Investigation, Study, Assay
- LIMS: laboratory information management system
- NFDI: German National Research Data Infrastructure
- NGS: next-generation sequencing
- OMIM: Online Mendelian Inheritance in Man
- PEP: Portable Encapsulated Projects
- RBAC: role-based access control
- RDM: resource data management
- SDM: scientific data management
- SDMS: scientific data management system
- SODAR: System for Omics Access and Retrieval
- TSV: tabular separated values
- VCF: variant call format
Acknowledgements
The authors thank all internal users, particularly CUBI members, for their feedback. Some icons from OpenMoji.org were used in the figures.
Author contributions
Conceptualization: M.N., M.H., D.B. Funding acquisition: D.B. Methodology: M.H., M.N. Project administration and supervision: M.H., D.B. Resources: D.B. Software: M.N., M.H., O.S., P.P. Writing and editing: all authors.
Funding
This work has been supported by the Ministry of Education and Research (BMBF), as part of the National Research Initiative “Mass Spectrometry in Systems Medicine” (MSCoreSys), grant-ID 031L0220A (MSTARS) and by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—project-ID 427826188–SFB 1444.
Data availability
All supporting data and materials are available in the GigaScience GigaDB database.[24]
Competing interests
The authors declare that they have no competing interests.
References
- ↑ Gonzalez, Andrew; Peres-Neto, Pedro R. (23 April 2015). "Act to staunch loss of research data" (in en). Nature 520 (7548): 436–436. doi:10.1038/520436c. ISSN 0028-0836. https://www.nature.com/articles/520436c.
- ↑ Wilkinson, Mark D.; Dumontier, Michel; Aalbersberg, IJsbrand Jan; Appleton, Gabrielle; Axton, Myles; Baak, Arie; Blomberg, Niklas; Boiten, Jan-Willem et al. (15 March 2016). "The FAIR Guiding Principles for scientific data management and stewardship" (in en). Scientific Data 3 (1): 160018. doi:10.1038/sdata.2016.18. ISSN 2052-4463. PMC PMC4792175. PMID 26978244. https://www.nature.com/articles/sdata201618.
- ↑ Donner, Eva Katharina (1 June 2023). "Research data management systems and the organization of universities and research institutes: A systematic literature review" (in en). Journal of Librarianship and Information Science 55 (2): 261–281. doi:10.1177/09610006211070282. ISSN 0961-0006. http://journals.sagepub.com/doi/10.1177/09610006211070282.
- ↑ 4.0 4.1 4.2 Machina, Hari K.; Wild, David J. (1 August 2013). "Electronic Laboratory Notebooks Progress and Challenges in Implementation" (in en). SLAS Technology 18 (4): 264–268. doi:10.1177/2211068213484471. https://linkinghub.elsevier.com/retrieve/pii/S2472630322016284.
- ↑ 5.0 5.1 Wolstencroft, Katherine; Owen, Stuart; Krebs, Olga; Nguyen, Quyen; Stanford, Natalie J; Golebiewski, Martin; Weidemann, Andreas; Bittkowski, Meik et al. (1 December 2015). "SEEK: a systems biology data and model management platform" (in en). BMC Systems Biology 9 (1): 33. doi:10.1186/s12918-015-0174-y. ISSN 1752-0509. PMC PMC4702362. PMID 26160520. https://bmcsystbiol.biomedcentral.com/articles/10.1186/s12918-015-0174-y.
- ↑ 6.0 6.1 King, Gary (1 November 2007). "An Introduction to the Dataverse Network as an Infrastructure for Data Sharing" (in en). Sociological Methods & Research 36 (2): 173–199. doi:10.1177/0049124107306660. ISSN 0049-1241. http://journals.sagepub.com/doi/10.1177/0049124107306660.
- ↑ 7.0 7.1 Smeele, T.; Westerhof, L. (2018). "Using iRODS to Manage, Share and Publish Research Data: Yoda" (PDF). iRODS User Group Meeting 2018 Proceedings: 5–12. https://irods.org/uploads/2018/irods_ugm2018_proceedings.pdf.
- ↑ Tryka, Kimberly A.; Hao, Luning; Sturcke, Anne; Jin, Yumi; Wang, Zhen Y.; Ziyabari, Lora; Lee, Moira; Popova, Natalia et al. (1 January 2014). "NCBI’s Database of Genotypes and Phenotypes: dbGaP" (in en). Nucleic Acids Research 42 (D1): D975–D979. doi:10.1093/nar/gkt1211. ISSN 0305-1048. PMC PMC3965052. PMID 24297256. https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gkt1211.
- ↑ Haug, Kenneth; Cochrane, Keeva; Nainala, Venkata Chandrasekhar; Williams, Mark; Chang, Jiakang; Jayaseelan, Kalai Vanii; O'Donovan, Claire (8 January 2020). "MetaboLights: a resource evolving in response to the needs of its scientific community". Nucleic Acids Research 48 (D1): D440–D444. doi:10.1093/nar/gkz1019. ISSN 1362-4962. PMC 7145518. PMID 31691833. https://pubmed.ncbi.nlm.nih.gov/31691833.
- ↑ 10.0 10.1 Mathé, Ewy; Davis, Sean, eds. (2016) (in en). Statistical Genomics: Methods and Protocols. Methods in Molecular Biology. 1418. New York, NY: Springer New York. doi:10.1007/978-1-4939-3578-9. ISBN 978-1-4939-3576-5. http://link.springer.com/10.1007/978-1-4939-3578-9.
- ↑ 11.0 11.1 van der Velde, K Joeri; Imhann, Floris; Charbon, Bart; Pang, Chao; van Enckevort, David; Slofstra, Mariska; Barbieri, Ruggero; Alberts, Rudi et al. (15 March 2019). Wren, Jonathan. ed. "MOLGENIS research: advanced bioinformatics data software for non-bioinformaticians" (in en). Bioinformatics 35 (6): 1076–1078. doi:10.1093/bioinformatics/bty742. ISSN 1367-4803. PMC PMC6419911. PMID 30165396. https://academic.oup.com/bioinformatics/article/35/6/1076/5085379.
- ↑ 12.0 12.1 Forschungszentrum Jülich; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad. "Zendro Documentation". GitHub. https://zendro-dev.github.io/. Retrieved 07 March 2023.
- ↑ 13.0 13.1 Sansone, Susanna-Assunta; Rocca-Serra, Philippe; Field, Dawn; Maguire, Eamonn; Taylor, Chris; Hofmann, Oliver; Fang, Hong; Neumann, Steffen et al. (1 February 2012). "Toward interoperable bioscience data" (in en). Nature Genetics 44 (2): 121–126. doi:10.1038/ng.1054. ISSN 1061-4036. PMC PMC3428019. PMID 22281772. https://www.nature.com/articles/ng.1054.
- ↑ Sheffield, Nathan C; Stolarczyk, Michał; Reuter, Vincent P; Rendeiro, André F (6 December 2021). "Linking big biomedical datasets to modular analysis with Portable Encapsulated Projects" (in en). GigaScience 10 (12): giab077. doi:10.1093/gigascience/giab077. ISSN 2047-217X. PMC PMC8673555. PMID 34890448. https://academic.oup.com/gigascience/article/doi/10.1093/gigascience/giab077/6454632.
- ↑ Gorgolewski, Krzysztof J.; Auer, Tibor; Calhoun, Vince D.; Craddock, R. Cameron; Das, Samir; Duff, Eugene P.; Flandin, Guillaume; Ghosh, Satrajit S. et al. (21 June 2016). "The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments" (in en). Scientific Data 3 (1): 160044. doi:10.1038/sdata.2016.44. ISSN 2052-4463. PMC PMC4978148. PMID 27326542. https://www.nature.com/articles/sdata201644.
- ↑ Facile, Rhonda; Muhlbradt, Erin Elizabeth; Gong, Mengchun; Li, Qingna; Popat, Vaishali; Pétavy, Frank; Cornet, Ronald; Ruan, Yaoping et al. (27 January 2022). "Use of Clinical Data Interchange Standards Consortium (CDISC) Standards for Real-world Data: Expert Perspectives From a Qualitative Delphi Survey" (in en). JMIR Medical Informatics 10 (1): e30363. doi:10.2196/30363. ISSN 2291-9694. PMC PMC8832264. PMID 35084343. https://medinform.jmir.org/2022/1/e30363.
- ↑ "The HDF5 Library and File Format". The HDF Group. https://www.hdfgroup.org/solutions/hdf5/. Retrieved 21 March 2023.
- ↑ Bischof, Jared; Wilke, Andreas; Gerlach, Wolfgang; Harrison, Travis; Paczian, Tobias; Tang, Wei; Trimble, William; Wilkening, Jared et al. (1 December 2015). "Shock: Active Storage for Multicloud Streaming Data Analysis". 2015 IEEE/ACM 2nd International Symposium on Big Data Computing (BDC) (Limassol: IEEE): 68–72. doi:10.1109/BDC.2015.40. ISBN 978-0-7695-5696-3. http://ieeexplore.ieee.org/document/7406331/.
- ↑ Ernst, M.; Fuhrmann, P.; Gasthuber, M. et al. (2001). Chen, H.S.. ed. "dCache, a distributed storage data caching system". Proceedings of CHEP 2001: 241–44. https://www.osti.gov/etdeweb/biblio/20408431.
- ↑ Hedges, Mark; Blanke, Tobias; Hasan, Adil (1 April 2009). "Rule-based curation and preservation of data: A data grid approach using iRODS" (in en). Future Generation Computer Systems 25 (4): 446–452. doi:10.1016/j.future.2008.10.003. https://linkinghub.elsevier.com/retrieve/pii/S0167739X08001660.
- ↑ Chiang, Gen-Tao; Clapham, Peter; Qi, Guoying; Sale, Kevin; Coates, Guy (1 December 2011). "Implementing a genomic data management system using iRODS in the Wellcome Trust Sanger Institute" (in en). BMC Bioinformatics 12 (1): 361. doi:10.1186/1471-2105-12-361. ISSN 1471-2105. PMC PMC3228552. PMID 21906284. https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-12-361.
- ↑ Courtot, Mélanie; Gupta, Dipayan; Liyanage, Isuru; Xu, Fuqi; Burdett, Tony (7 January 2022). "BioSamples database: FAIRer samples metadata to accelerate research data management" (in en). Nucleic Acids Research 50 (D1): D1500–D1507. doi:10.1093/nar/gkab1046. ISSN 0305-1048. PMC PMC8728232. PMID 34747489. https://academic.oup.com/nar/article/50/D1/D1500/6423179.
- ↑ Leinonen, R.; Sugawara, H.; Shumway, M.; on behalf of the International Nucleotide Sequence Database Collaboration (1 January 2011). "The Sequence Read Archive" (in en). Nucleic Acids Research 39 (Database): D19–D21. doi:10.1093/nar/gkq1019. ISSN 0305-1048. PMC PMC3013647. PMID 21062823. https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gkq1019.
- ↑ 24.0 24.1 24.2 Mikko, Nieminen; Oliver, Stolpe; Mathias, Kuhring; January, Weiner; Patrick, Pett; Dieter, Beule; Manuel, Holtgrewe (2023), "Supporting data for "SODAR: managing multi-omics study data and metadata"" (in en), GigaDB (GigaScience Database), doi:10.5524/102401, http://gigadb.org/dataset/102401. Retrieved 2024-01-02
- ↑ Mohr, Christopher; Friedrich, Andreas; Wojnar, David; Kenar, Erhan; Polatkan, Aydin Can; Codrea, Marius Cosmin; Czemmel, Stefan; Kohlbacher, Oliver et al. (19 January 2018). Lisacek, Frederique. ed. "qPortal: A platform for data-driven biomedical research" (in en). PLOS ONE 13 (1): e0191603. doi:10.1371/journal.pone.0191603. ISSN 1932-6203. PMC PMC5774839. PMID 29352322. https://dx.plos.org/10.1371/journal.pone.0191603.
- ↑ Barillari, Caterina; Ottoz, Diana S. M.; Fuentes-Serna, Juan Mariano; Ramakrishnan, Chandrasekhar; Rinn, Bernd; Rudolf, Fabian (15 February 2016). "openBIS ELN-LIMS: an open-source database for academic laboratories" (in en). Bioinformatics 32 (4): 638–640. doi:10.1093/bioinformatics/btv606. ISSN 1367-4811. PMC PMC4743625. PMID 26508761. https://academic.oup.com/bioinformatics/article/32/4/638/1743839.
- ↑ CARPi, Nicolas; Minges, Alexander; Piel, Matthieu (14 April 2017). "eLabFTW: An open source laboratory notebook for research labs". The Journal of Open Source Software 2 (12): 146. doi:10.21105/joss.00146. ISSN 2475-9066. http://joss.theoj.org/papers/10.21105/joss.00146.
- ↑ Kuhn, R. M.; Haussler, D.; Kent, W. J. (1 March 2013). "The UCSC genome browser and associated tools" (in en). Briefings in Bioinformatics 14 (2): 144–161. doi:10.1093/bib/bbs038. ISSN 1467-5463. PMC PMC3603215. PMID 22908213. https://academic.oup.com/bib/article-lookup/doi/10.1093/bib/bbs038.
- ↑ Obermayer, Benedikt; Holtgrewe, Manuel; Nieminen, Mikko; Messerschmidt, Clemens; Beule, Dieter (19 February 2020). "SCelVis: exploratory single cell data analysis on the desktop and in the cloud" (in en). PeerJ 8: e8607. doi:10.7717/peerj.8607. ISSN 2167-8359. PMC PMC7035868. PMID 32117635. https://peerj.com/articles/8607.
- ↑ Robinson, James T; Thorvaldsdóttir, Helga; Winckler, Wendy; Guttman, Mitchell; Lander, Eric S; Getz, Gad; Mesirov, Jill P (1 January 2011). "Integrative genomics viewer" (in en). Nature Biotechnology 29 (1): 24–26. doi:10.1038/nbt.1754. ISSN 1087-0156. PMC PMC3346182. PMID 21221095. https://www.nature.com/articles/nbt.1754.
- ↑ "The German Human Genome-Phenome Archive". Deutsche Forschungsgemeinschaft. https://www.ghga.de/. Retrieved 28 March 2023.
- ↑ Freeberg, Mallory Ann; Fromont, Lauren A; D’Altri, Teresa; Romero, Anna Foix; Ciges, Jorge Izquierdo; Jene, Aina; Kerry, Giselle; Moldes, Mauricio et al. (7 January 2022). "The European Genome-phenome Archive in 2021" (in en). Nucleic Acids Research 50 (D1): D980–D987. doi:10.1093/nar/gkab1059. ISSN 0305-1048. PMC PMC8728218. PMID 34791407. https://academic.oup.com/nar/article/50/D1/D980/6430505.
- ↑ Kuhring, Mathias; Nieminen, Mikko; Kirwan, Jennifer; Beule, Dieter; Holtgrewe, Manuel (20 August 2019). "AltamISA: a Python API for ISA-Tab files". Journal of Open Source Software 4 (40): 1610. doi:10.21105/joss.01610. ISSN 2475-9066. https://joss.theoj.org/papers/10.21105/joss.01610.
- ↑ Ferraiolo, David; Kuhn, D. Richard; Chandramouli, Ramaswamy (2007). Role-based access control. Artech House information security and privacy series (2nd ed ed.). Boston: Artech House. ISBN 978-1-59693-113-8. OCLC ocm85851304. https://www.worldcat.org/title/mediawiki/oclc/ocm85851304.
- ↑ Smeele, T.; Smeele, C. (2016). "Davrods, an Apache WebDAV Interface to iRODS" (PDF). iRODS User Group Meeting 2016 Proceedings: 41–48. https://irods.org/uploads/2016/12/irods_ugm2016_proceedings.pdf.
- ↑ Köhler, Sebastian; Gargano, Michael; Matentzoglu, Nicolas; Carmody, Leigh C; Lewis-Smith, David; Vasilevsky, Nicole A; Danis, Daniel; Balagura, Ganna et al. (8 January 2021). "The Human Phenotype Ontology in 2021" (in en). Nucleic Acids Research 49 (D1): D1207–D1217. doi:10.1093/nar/gkaa1043. ISSN 0305-1048. PMC PMC7778952. PMID 33264411. https://academic.oup.com/nar/article/49/D1/D1207/6017351.
- ↑ Hamosh, A.; Scott, A.F.; Amberger, J.S. et al. (17 December 2004). "Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders" (in en). Nucleic Acids Research 33 (Database issue): D514–D517. doi:10.1093/nar/gki033. ISSN 1362-4962. PMC PMC539987. PMID 15608251. https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gki033.
- ↑ Federhen, S. (1 January 2012). "The NCBI Taxonomy database" (in en). Nucleic Acids Research 40 (D1): D136–D143. doi:10.1093/nar/gkr1178. ISSN 0305-1048. PMC PMC3245000. PMID 22139910. https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gkr1178.
- ↑ Nieminen, Mikko; Stolpe, Oliver; Schumann, Franziska; Holtgrewe, Manuel; Beule, Dieter (15 November 2020). "SODAR Core: a Django-based framework for scientific data management and analysis web apps". Journal of Open Source Software 5 (55): 1584. doi:10.21105/joss.01584. ISSN 2475-9066. https://joss.theoj.org/papers/10.21105/joss.01584.
- ↑ Holtgrewe, Manuel; Stolpe, Oliver; Nieminen, Mikko; Mundlos, Stefan; Knaus, Alexej; Kornak, Uwe; Seelow, Dominik; Segebrecht, Lara et al. (2 July 2020). "VarFish: comprehensive DNA variant analysis for diagnostics and research" (in en). Nucleic Acids Research 48 (W1): W162–W169. doi:10.1093/nar/gkaa241. ISSN 0305-1048. PMC PMC7319464. PMID 32338743. https://academic.oup.com/nar/article/48/W1/W162/5825625.
- ↑ Stolpe, O.; Nieminen, M.; Holtgrewe, M. et al.. "bihealth / kiosc-server". GitHub. https://github.com/bihealth/kiosc-server. Retrieved 28 June 2023.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, grammar, and punctuation. In some cases important information was missing from the references, and that information was added.