Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Grimes BMCBioinformatics2014 15.jpg|220px]]</div> | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Grimes BMCBioinformatics2014 15.jpg|220px]]</div> | ||
'''"[[Journal: | '''"[[Journal:adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies|adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies]]"''' | ||
Many biological laboratories that deal with genomic samples are facing the problem of sample tracking, both for pure [[laboratory]] management and for efficiency. Our laboratory exploits PCR techniques and Next Generation Sequencing (NGS) methods to perform high-throughput integration site monitoring in different clinical trials and scientific projects. Because of the huge amount of samples that we process every year, which result in hundreds of millions of sequencing reads, we need to standardize data management and tracking systems, building up a scalable and flexible structure with web-based interfaces, which are usually called [[Laboratory information management system|Laboratory Information Management System]] (LIMS). | |||
We extended and customized ADempiere ERP to fulfill LIMS requirements and we developed adLIMS. It has been validated by our end-users verifying functionalities and GUIs through test cases for PCRs samples and pre-sequencing data and it is currently in use in our laboratories. adLIMS implements authorization and authentication policies, allowing multiple users management and roles definition that enables specific permissions, operations and data views to each user. ('''[[Journal:adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
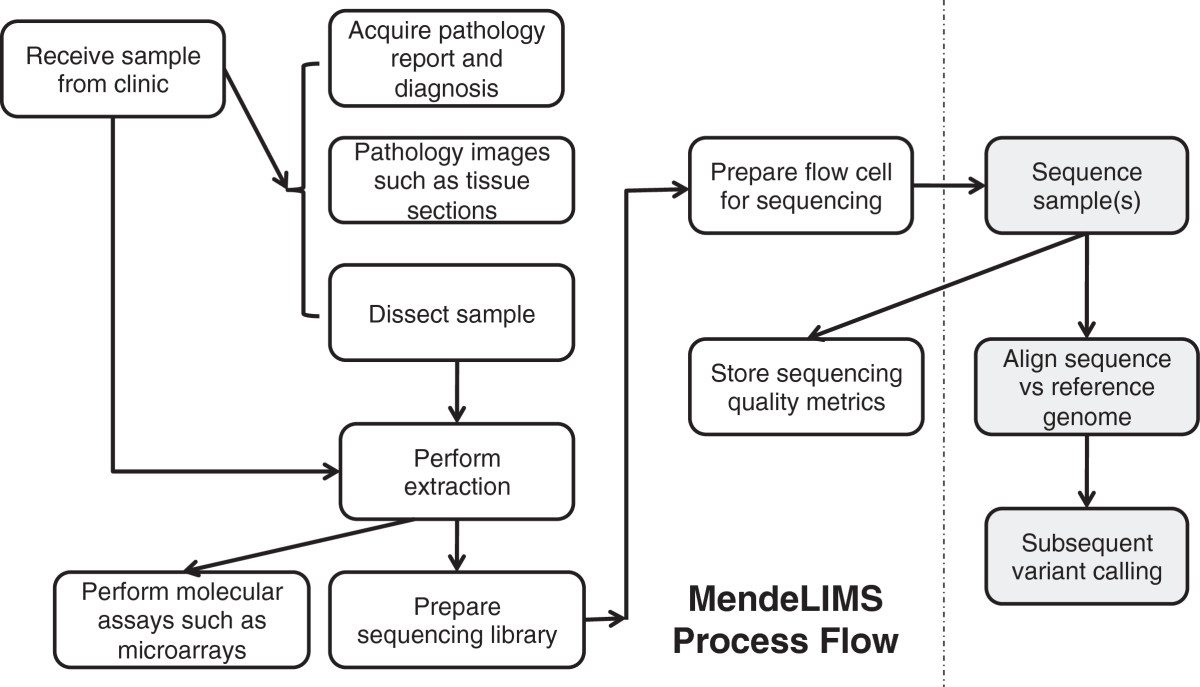
''Recently featured'': [[Journal:Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages|Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages]], [[Journal:Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations|Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations | ''Recently featured'': [[Journal:MendeLIMS: A web-based laboratory information management system for clinical genome sequencing|MendeLIMS: A web-based laboratory information management system for clinical genome sequencing]], [[Journal:Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages|Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages]], [[Journal:Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations|Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations]] | ||
Revision as of 20:00, 11 January 2016
Many biological laboratories that deal with genomic samples are facing the problem of sample tracking, both for pure laboratory management and for efficiency. Our laboratory exploits PCR techniques and Next Generation Sequencing (NGS) methods to perform high-throughput integration site monitoring in different clinical trials and scientific projects. Because of the huge amount of samples that we process every year, which result in hundreds of millions of sequencing reads, we need to standardize data management and tracking systems, building up a scalable and flexible structure with web-based interfaces, which are usually called Laboratory Information Management System (LIMS).
We extended and customized ADempiere ERP to fulfill LIMS requirements and we developed adLIMS. It has been validated by our end-users verifying functionalities and GUIs through test cases for PCRs samples and pre-sequencing data and it is currently in use in our laboratories. adLIMS implements authorization and authentication policies, allowing multiple users management and roles definition that enables specific permissions, operations and data views to each user. (Full article...)
Recently featured: MendeLIMS: A web-based laboratory information management system for clinical genome sequencing, Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages, Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations