Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Garza BMCBioinformatics2016 17.gif|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:From the desktop to the grid: Scalable bioinformatics via workflow conversion|From the desktop to the grid: Scalable bioinformatics via workflow conversion]]"''' | ||
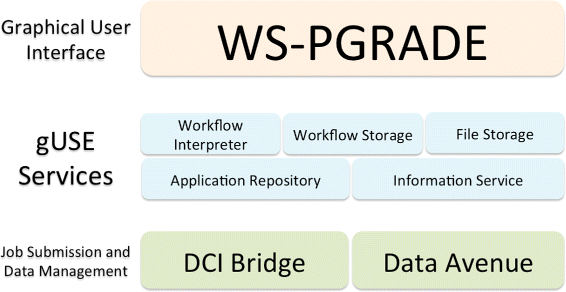
Reproducibility is one of the tenets of the [[scientific method]]. Scientific experiments often comprise complex data flows, selection of adequate parameters, and analysis and visualization of intermediate and end results. Breaking down the complexity of such experiments into the joint collaboration of small, repeatable, well defined tasks, each with well defined inputs, parameters, and outputs, offers the immediate benefit of identifying bottlenecks, pinpoint sections which could benefit from parallelization, among others. [[Workflow]]s rest upon the notion of splitting complex work into the joint effort of several manageable tasks. | |||
There are several engines that give users the ability to design and execute workflows. Each engine was created to address certain problems of a specific community, therefore each one has its advantages and shortcomings. Furthermore, not all features of all workflow engines are royalty-free — an aspect that could potentially drive away members of the scientific community. ('''[[Journal:From the desktop to the grid: Scalable bioinformatics via workflow conversion|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Terminology spectrum analysis of natural-language chemical documents: Term-like phrases retrieval routine|Terminology spectrum analysis of natural-language chemical documents: Term-like phrases retrieval routine]] | |||
: ▪ [[Journal:A legal framework to support development and assessment of digital health services|A legal framework to support development and assessment of digital health services]] | : ▪ [[Journal:A legal framework to support development and assessment of digital health services|A legal framework to support development and assessment of digital health services]] | ||
: ▪ [[Journal:The GAAIN Entity Mapper: An active-learning system for medical data mapping|The GAAIN Entity Mapper: An active-learning system for medical data mapping]] | : ▪ [[Journal:The GAAIN Entity Mapper: An active-learning system for medical data mapping|The GAAIN Entity Mapper: An active-learning system for medical data mapping]] | ||
Revision as of 15:42, 15 August 2016
"From the desktop to the grid: Scalable bioinformatics via workflow conversion"
Reproducibility is one of the tenets of the scientific method. Scientific experiments often comprise complex data flows, selection of adequate parameters, and analysis and visualization of intermediate and end results. Breaking down the complexity of such experiments into the joint collaboration of small, repeatable, well defined tasks, each with well defined inputs, parameters, and outputs, offers the immediate benefit of identifying bottlenecks, pinpoint sections which could benefit from parallelization, among others. Workflows rest upon the notion of splitting complex work into the joint effort of several manageable tasks.
There are several engines that give users the ability to design and execute workflows. Each engine was created to address certain problems of a specific community, therefore each one has its advantages and shortcomings. Furthermore, not all features of all workflow engines are royalty-free — an aspect that could potentially drive away members of the scientific community. (Full article...)
Recently featured: