Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Barker IntJourSTEMEd2015 2.jpg|220px]]</div> | ||
'''"[[Journal: | '''"[[Journal:University-level practical activities in bioinformatics benefit voluntary groups of pupils in the last 2 years of school|University-level practical activities in bioinformatics benefit voluntary groups of pupils in the last 2 years of school]]"''' | ||
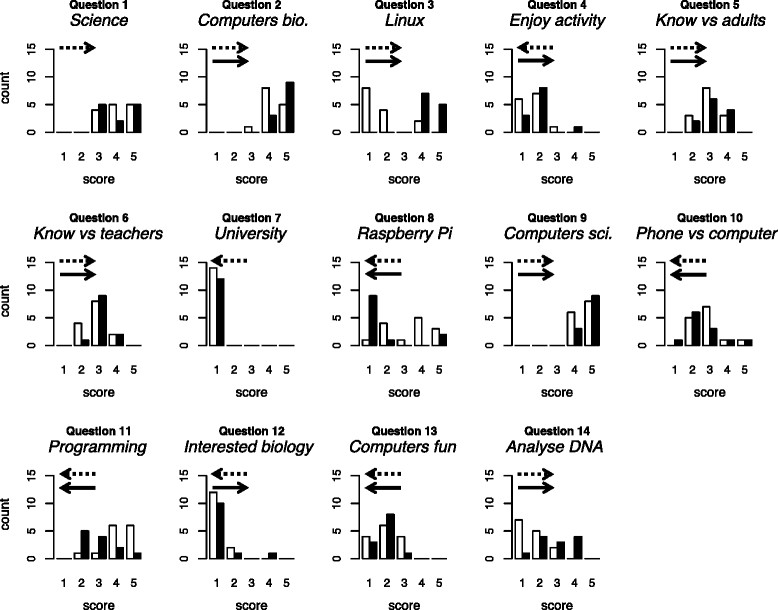
[[Bioinformatics]] — the use of computers in biology — is of major and increasing importance to biological sciences and medicine. We conducted a preliminary investigation of the value of bringing practical, university-level bioinformatics education to the school level. We conducted voluntary activities for pupils at two schools in Scotland (years S5 and S6; pupils aged 15–17). We used material originally developed for an optional final-year undergraduate module and now incorporated into 4273''π'', a resource for teaching and learning bioinformatics on the low-cost Raspberry Pi computer. | |||
Pupils’ feedback forms suggested our activities were beneficial. During the course of the activity, they provide strong evidence of increase in the following: pupils’ perception of the value of computers within biology; their knowledge of the Linux operating system and the Raspberry Pi; their willingness to use computers rather than phones or tablets; their ability to program a computer and their ability to analyse DNA sequences with a computer. We found no strong evidence of negative effects. | |||
Our preliminary study supports the feasibility of bringing university-level, practical bioinformatics activities to school pupils. ('''[[Journal:University-level practical activities in bioinformatics benefit voluntary groups of pupils in the last 2 years of school|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': [[Journal:Factors associated with adoption of health information technology: A conceptual model based on a systematic review|Factors associated with adoption of health information technology: A conceptual model based on a systematic review]][[Journal:Generalized procedure for screening free software and open-source software applications|Generalized procedure for screening free software and open-source software applications | ''Recently featured'': [[Journal:Support patient search on pathology reports with interactive online learning based data extraction|Support patient search on pathology reports with interactive online learning based data extraction]], [[Journal:Factors associated with adoption of health information technology: A conceptual model based on a systematic review|Factors associated with adoption of health information technology: A conceptual model based on a systematic review]][[Journal:Generalized procedure for screening free software and open-source software applications|Generalized procedure for screening free software and open-source software applications]] | ||
Revision as of 18:12, 30 November 2015
Bioinformatics — the use of computers in biology — is of major and increasing importance to biological sciences and medicine. We conducted a preliminary investigation of the value of bringing practical, university-level bioinformatics education to the school level. We conducted voluntary activities for pupils at two schools in Scotland (years S5 and S6; pupils aged 15–17). We used material originally developed for an optional final-year undergraduate module and now incorporated into 4273π, a resource for teaching and learning bioinformatics on the low-cost Raspberry Pi computer.
Pupils’ feedback forms suggested our activities were beneficial. During the course of the activity, they provide strong evidence of increase in the following: pupils’ perception of the value of computers within biology; their knowledge of the Linux operating system and the Raspberry Pi; their willingness to use computers rather than phones or tablets; their ability to program a computer and their ability to analyse DNA sequences with a computer. We found no strong evidence of negative effects.
Our preliminary study supports the feasibility of bringing university-level, practical bioinformatics activities to school pupils. (Full article...)
Recently featured: Support patient search on pathology reports with interactive online learning based data extraction, Factors associated with adoption of health information technology: A conceptual model based on a systematic reviewGeneralized procedure for screening free software and open-source software applications