Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Reid BMCInformatics2014 15.jpg|220px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Launching genomics into the cloud: Deployment of Mercury, a next generation sequence analysis pipeline|Launching genomics into the cloud: Deployment of Mercury, a next generation sequence analysis pipeline]]"''' | ||
Massively parallel DNA sequencing generates staggering amounts of data. Decreasing cost, increasing throughput, and improved annotation have expanded the diversity of genomics applications in research and clinical practice. This expanding scale creates analytical challenges: accommodating peak compute demand, coordinating secure access for multiple analysts, and sharing validated tools and results. | |||
To address these challenges, we have developed the Mercury analysis pipeline and deployed it in local hardware and the Amazon Web Services cloud via the DNAnexus platform. Mercury is an automated, flexible, and extensible analysis workflow that provides accurate and reproducible genomic results at scales ranging from individuals to large cohorts. | |||
By taking advantage of cloud computing and with Mercury implemented on the DNAnexus platform, we have demonstrated a powerful combination of a robust and fully validated software pipeline and a scalable computational resource that, to date, we have applied to more than 10,000 whole genome and whole exome samples. ('''[[Journal:Launching genomics into the cloud: Deployment of Mercury, a next generation sequence analysis pipeline|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': [[Journal:adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies|adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies]], [[Journal:MendeLIMS: A web-based laboratory information management system for clinical genome sequencing|MendeLIMS: A web-based laboratory information management system for clinical genome sequencing | ''Recently featured'': [[Journal:Benefits of the community for partners of open source vendors|Benefits of the community for partners of open source vendors]], [[Journal:adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies|adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies]], [[Journal:MendeLIMS: A web-based laboratory information management system for clinical genome sequencing|MendeLIMS: A web-based laboratory information management system for clinical genome sequencing]] | ||
Revision as of 16:46, 25 January 2016
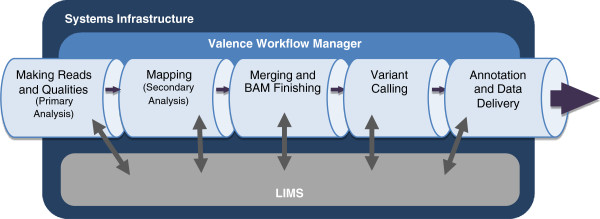
Massively parallel DNA sequencing generates staggering amounts of data. Decreasing cost, increasing throughput, and improved annotation have expanded the diversity of genomics applications in research and clinical practice. This expanding scale creates analytical challenges: accommodating peak compute demand, coordinating secure access for multiple analysts, and sharing validated tools and results.
To address these challenges, we have developed the Mercury analysis pipeline and deployed it in local hardware and the Amazon Web Services cloud via the DNAnexus platform. Mercury is an automated, flexible, and extensible analysis workflow that provides accurate and reproducible genomic results at scales ranging from individuals to large cohorts.
By taking advantage of cloud computing and with Mercury implemented on the DNAnexus platform, we have demonstrated a powerful combination of a robust and fully validated software pipeline and a scalable computational resource that, to date, we have applied to more than 10,000 whole genome and whole exome samples. (Full article...)
Recently featured: Benefits of the community for partners of open source vendors, adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies, MendeLIMS: A web-based laboratory information management system for clinical genome sequencing