Difference between revisions of "User:Shawndouglas/sandbox/sublevel1"
Shawndouglas (talk | contribs) |
Shawndouglas (talk | contribs) |
||
| Line 1: | Line 1: | ||
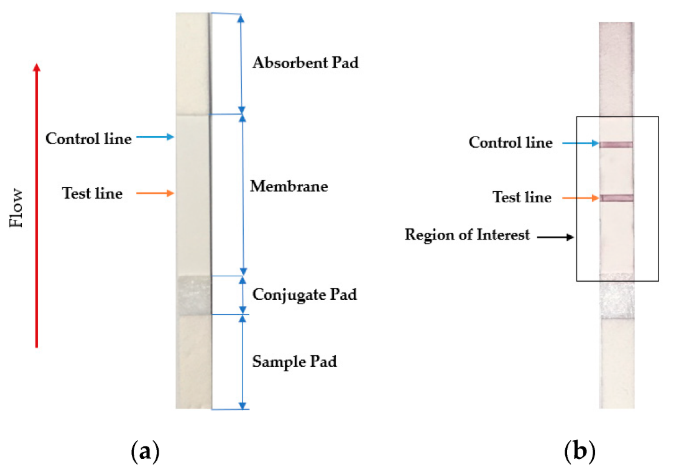
[[File:Fig2 Foysal Sensors2019 19-21.png|right|thumb|430px|('''a''') Sample LFA strip and ('''b''') region of interest of an LFA strip with an analyte. The intensity and density of red color in the test line region determine the amount of analyte in the sample. ([https://doi.org/10.3390/s19214812 From Foysal ''et al.'' 2019])]]The lateral flow assay or LFA is another molecular biology method for detecting the presence of a target analyte in specimen material. In comparison to PCR, LFA has the advantages of being more rapid, low-cost, easy-to-use, and applicable at the point of care, though the average LFA is at best semi-quantitative in its results and has slightly lower sensitivity.<ref name="KoczulaLateral16">{{cite journal |title=Lateral flow assays |journal=Essays in Biochemistry |author=Koczula, K.M.; Gallotta, A. |volume=60 |issue=1 |pages=111–20 |year=2016 |doi=10.1042/EBC20150012 |pmid=27365041 |pmc=PMC4986465}}</ref><ref name="LiuPaper18">{{cite book |url=https://books.google.com/books?id=jSk0DwAAQBAJ&pg=PA183 |chapter=Chapter 8: Paper-Based Immunoassays |title=Handbook of Immunoassay Technologies: Approaches, Performances, and Applications |author=Liu, Z.; Hu, J.; Qu, Z.; Xu, F. |editor=Vashist, S.K.; Luong, J.H.T. |publisher=Academic Press |pages=183–202 |year=2018 |isbn=9780128117620 |accessdate=08 April 2020}}</ref> The method involves a cellulose-based strip with an ordered collection of specific "pads" and reagents that are reactive to a target analyte in a liquid, which is placed on the strip and moves across the various reagents using capillary and electrostatic interactions.<ref name="KoczulaLateral16" /><ref name="Jauset-RubioUltra16">{{cite journal |title=Ultrasensitive, rapid and inexpensive detection of DNA using paper based lateral flow assay |journal=Scientific Reports |author=Jauset-Rubio, M.; Svobodová, M.; Mairal, T. et al. |volume=6 |at=37732 |year=2016 |doi=10.1038/srep37732 |pmid=27886248 |pmc=PMC5123575}}</ref><ref name="FoysalAnalyte19">{{cite journal |title=Analyte Quantity Detection from Lateral Flow Assay Using a Smartphone |journal=Sensors |author=Foysal, K.H.; Seo, S.E.; Kim, M.J. et al. |volume=19 |issue=21 |at=4812 |year=2019 |doi=10.3390/s19214812 |pmid=31694281 |pmc=PMC6864604}}</ref> LFA has been used in molecular diagnostics for testing urine, saliva, sweat, serum, plasma, and other biological fluids for the presence of specific antigens and antibodies, as well as signs of gene amplification. The method has also been effectively applied to other industries outside healthcare, including the food and beverage, chemical, and environmental industries.<ref name="KoczulaLateral16" /> | |||
In the realm of infectious disease, the LFA has played an important role in disease diagnosis and control, particularly in resource-constrained settings where resources are limited and point-of-care testing is critical to filling the gap.<ref name="HanafiahDevelop17">{{cite journal |title=Development of Multiplexed Infectious Disease Lateral Flow Assays: Challenges and Opportunities |journal=Diagnostics |author=Hanafiah, K.M.; Arifin, N.; Bustami, T. et al. |volume=7 |issue=3 |at=51 |year=2017 |doi=10.3390/diagnostics7030051 |pmid=28880218 |pmc=PMC5617951}}</ref> Testing for the presence of an infectious agent in body fluids using LFA can be performed in two ways: lateral flow immunoassay (LFI) or lateral flow nucleic acid (LFNA). The immunoassay variant looks for antibodies created as a result of the presence of an infectious agent, whereas the nucleic acid variant is built to detect an amplified nucleic acid sequence specific to a target infectious agent.<ref name="NgomDevelop10">{{cite journal |title=Development and application of lateral flow test strip technology for detection of infectious agents and chemical contaminants: A review |journal=Analytical and Bioanalytical Chemistry |author=Ngom, B.; Guo, Y.; Wang, X.; Bi, D. |volume=397 |pages=1113–1135 |year=2010 |doi=10.1007/s00216-010-3661-4 |pmid=20422164}}</ref> Speaking specifically to the SARS-CoV-2 coronavirus, the antibody-based immunoassay method of lateral flow is predominantly used, targeting one of either a monoclonal antibody directed at a viral antigen, or a viral antigen that is recognizable by a patient's developed antibodies.<ref name="SheridanFast20">{{cite web |url=https://www.nature.com/articles/d41587-020-00010-2 |title=Fast, portable tests come online to curb coronavirus pandemic |author=Sheridan, C. |work=Nature Biotechnology - News |date=23 March 2020 |accessdate=08 April 2020 |doi=10.1038/d41587-020-00010-2}}</ref><ref name="RBCoronavirus20">{{cite web |url=https://www.raybiotech.com/index.php?dispatch=attachments.getfile&attachment_id=3095 |archiveurl=https://web.archive.org/web/20200408211820/https://www.raybiotech.com/index.php?dispatch=attachments.getfile&attachment_id=3095 |format=PDF |title=Novel Coronavirus (SARS-CoV-2) IgM Antibody Detection Kit (Colloidal Gold Method) |publisher=RayBiotech |date=03 April 2020 |archivedate=08 April 2020 |accessdate=06 September 2021}}</ref> | |||
Testing using LFI is generally as follows. A specialized adsorbent sample pad at one end of the LFI strip receives the specimen material. That material then migrates to the next conjugate release pad, where the specimen material is exposed to "antibodies that are specific to the target analyte and are conjugated to coloured or fluorescent particles."<ref name="KoczulaLateral16" /> The material then progresses to a detection zone containing antibodies or antigens that are fixed in the zone and intended to react to a specific analyte. If the analyte is present, a test line produces a visual, qualitative response, and a control line ensures proper liquid flow across the strip. A wicking pad at the other end properly maintains the flow of liquids across the strip.<ref name="KoczulaLateral16" /> | |||
==References== | ==References== | ||
{{Reflist|colwidth=30em}} | {{Reflist|colwidth=30em}} | ||
Revision as of 16:46, 3 February 2022
The lateral flow assay or LFA is another molecular biology method for detecting the presence of a target analyte in specimen material. In comparison to PCR, LFA has the advantages of being more rapid, low-cost, easy-to-use, and applicable at the point of care, though the average LFA is at best semi-quantitative in its results and has slightly lower sensitivity.[1][2] The method involves a cellulose-based strip with an ordered collection of specific "pads" and reagents that are reactive to a target analyte in a liquid, which is placed on the strip and moves across the various reagents using capillary and electrostatic interactions.[1][3][4] LFA has been used in molecular diagnostics for testing urine, saliva, sweat, serum, plasma, and other biological fluids for the presence of specific antigens and antibodies, as well as signs of gene amplification. The method has also been effectively applied to other industries outside healthcare, including the food and beverage, chemical, and environmental industries.[1]
In the realm of infectious disease, the LFA has played an important role in disease diagnosis and control, particularly in resource-constrained settings where resources are limited and point-of-care testing is critical to filling the gap.[5] Testing for the presence of an infectious agent in body fluids using LFA can be performed in two ways: lateral flow immunoassay (LFI) or lateral flow nucleic acid (LFNA). The immunoassay variant looks for antibodies created as a result of the presence of an infectious agent, whereas the nucleic acid variant is built to detect an amplified nucleic acid sequence specific to a target infectious agent.[6] Speaking specifically to the SARS-CoV-2 coronavirus, the antibody-based immunoassay method of lateral flow is predominantly used, targeting one of either a monoclonal antibody directed at a viral antigen, or a viral antigen that is recognizable by a patient's developed antibodies.[7][8]
Testing using LFI is generally as follows. A specialized adsorbent sample pad at one end of the LFI strip receives the specimen material. That material then migrates to the next conjugate release pad, where the specimen material is exposed to "antibodies that are specific to the target analyte and are conjugated to coloured or fluorescent particles."[1] The material then progresses to a detection zone containing antibodies or antigens that are fixed in the zone and intended to react to a specific analyte. If the analyte is present, a test line produces a visual, qualitative response, and a control line ensures proper liquid flow across the strip. A wicking pad at the other end properly maintains the flow of liquids across the strip.[1]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Koczula, K.M.; Gallotta, A. (2016). "Lateral flow assays". Essays in Biochemistry 60 (1): 111–20. doi:10.1042/EBC20150012. PMC PMC4986465. PMID 27365041. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4986465.
- ↑ Liu, Z.; Hu, J.; Qu, Z.; Xu, F. (2018). "Chapter 8: Paper-Based Immunoassays". In Vashist, S.K.; Luong, J.H.T.. Handbook of Immunoassay Technologies: Approaches, Performances, and Applications. Academic Press. pp. 183–202. ISBN 9780128117620. https://books.google.com/books?id=jSk0DwAAQBAJ&pg=PA183. Retrieved 08 April 2020.
- ↑ Jauset-Rubio, M.; Svobodová, M.; Mairal, T. et al. (2016). "Ultrasensitive, rapid and inexpensive detection of DNA using paper based lateral flow assay". Scientific Reports 6: 37732. doi:10.1038/srep37732. PMC PMC5123575. PMID 27886248. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5123575.
- ↑ Foysal, K.H.; Seo, S.E.; Kim, M.J. et al. (2019). "Analyte Quantity Detection from Lateral Flow Assay Using a Smartphone". Sensors 19 (21): 4812. doi:10.3390/s19214812. PMC PMC6864604. PMID 31694281. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6864604.
- ↑ Hanafiah, K.M.; Arifin, N.; Bustami, T. et al. (2017). "Development of Multiplexed Infectious Disease Lateral Flow Assays: Challenges and Opportunities". Diagnostics 7 (3): 51. doi:10.3390/diagnostics7030051. PMC PMC5617951. PMID 28880218. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5617951.
- ↑ Ngom, B.; Guo, Y.; Wang, X.; Bi, D. (2010). "Development and application of lateral flow test strip technology for detection of infectious agents and chemical contaminants: A review". Analytical and Bioanalytical Chemistry 397: 1113–1135. doi:10.1007/s00216-010-3661-4. PMID 20422164.
- ↑ Sheridan, C. (23 March 2020). "Fast, portable tests come online to curb coronavirus pandemic". Nature Biotechnology - News. doi:10.1038/d41587-020-00010-2. https://www.nature.com/articles/d41587-020-00010-2. Retrieved 08 April 2020.









