Journal:SistematX, an online web-based cheminformatics tool for data management of secondary metabolites
| Full article title | SistematX, an online web-based cheminformatics tool for data management of secondary metabolites |
|---|---|
| Journal | Molecules |
| Author(s) |
Scotti, Marcus T.; Herrera-Acevedo, Chonny; Oliveira, Tiago B.; Costa, Renan P.O.; de Oliveira Santos, Silas Y.K.; Rodrigues, Ricardo P.; Scotti, Luciana; Da-Costa, Fernando B. |
| Author affiliation(s) | Federal University of Paraíba, Federal University of Sergipe, University of São Paulo |
| Primary contact | Phone: +55-83-99869-0415 |
| Year published | 2018 |
| Volume and issue | 23(1) |
| Page(s) | 103 |
| DOI | 10.3390/molecules23010103 |
| ISSN | 1420-3049 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://www.mdpi.com/1420-3049/23/1/103/htm |
| Download | http://www.mdpi.com/1420-3049/23/1/103/pdf (PDF) |
Abstract
The traditional work of a natural products researcher consists in large part of time-consuming experimental work, collecting biota to prepare, extracts to analyze, and innovative metabolites to identify. However, along this long scientific path, much information is lost or restricted to a specific niche. The large amounts of data already produced and the science of metabolomics reveal new questions: Are these compounds known or new? How fast can this information be obtained? To answer these and other relevant questions, an appropriate procedure to correctly store information on the data retrieved from the discovered metabolites is necessary. The SistematX (http://sistematx.ufpb.br) interface is implemented considering the following aspects: (a) the ability to search by structure, SMILES (Simplified Molecular-Input Line-Entry System) code, compound name, and species; (b) the ability to save chemical structures found by searching; (c) the ability to display compound data results, including important characteristics for natural products chemistry; and (d) the user's ability to find specific information for taxonomic rank (from family to species) of the plant from which the compound was isolated, the searched-for molecule, and the bibliographic reference and Global Positioning System (GPS) coordinates. The SistematX homepage allows the user to log into the data management area using a login name and password and gain access to administration pages. In this article, we introduce a modern and innovative web interface for the management of a secondary metabolite database. With its multi-platform design, it is able to be properly consulted via the internet and managed from any accredited computer. The interface provided by SistematX contains a wealth of useful information for the scientific community about natural products, highlighting the locations of species from which compounds are isolated.
Keywords: SistematX, secondary metabolites, data management, online web-based tool
Introduction
The traditional work of a natural products researcher can be summarized as the collection of biological samples, preparation of extracts for biological screening or bioassay-guided fractionation, and isolation and purification of (bioactive or not) compounds. However, the first question that may arise is the following: are these compounds known or new? In addition, metabolomics studies have introduced a new question: how fast can this information be obtained?[1]
The stage of dereplication, a process known as the rapid characterization of previously known compounds in mixtures without their prior purification, has become a strategically important area for natural products research involved in screening programs in several commercial and non-commercial databases.[2][3][4] These databases can be searched with minimal information, such as structural chemical and biological data from compounds; however, dereplication now requires additional information, such as biogeographical and taxonomic information, or the presence of a certain compound (new or known) in other individuals of the same species, genus, subfamily, and family. This information can also help to reduce the number of hits during chemical identification by dereplication.
Large structure-based data collections, such as ChemSpider[5], PubChem[6], ChEBI[7], and ZINC[8] can be used for this purpose.[9][10] However, these databases are not specialized in secondary metabolite information that is valuable to the natural products researchers, for example, botanical occurrence and geographical localization. For this reason, a number of specialized natural products databases were developed that are commercially or freely available and only contain restricted information, for example, the Dictionary of Natural Products (DNP)[11], NAPRALERT[12], Marinlit for marine natural products[13], and Antibase for microorganisms and higher fungi materials. Nevertheless, none of these provide structural collections in a format that can be rapidly integrated into software such as ACD/Structure Elucidator and others.[9]
Other natural products databases provide natural products extracted from various resources and contain various associated information such as toxicity prediction, but so far, little or nothing is known about these resources, for example, SUPER NATURAL II.[14] Natural products databases exhibit a huge range of structural complexity and thus are expected to contribute to the ability of such databases to provide positive hits.[2][15] These structures are available in regional databases, for example NeBBEDB[16], SANCDB[17], TM-CM[18], TCM-Database@Taiwan[19], NANPDB[20], and TCMID.[21] Many have been used in virtual screening research studies. In addition to the database information described above that uses two-dimensional (2D) structures, several databases have selected methods and tools for generating three-dimensional (3D) structures of small organic molecules, often for use in structure-based drug design.
In addition, databases of natural products with a focus on metabolomic studies with relationships between species-metabolites include the KNApSAcK Family[22], TIPdb-3D[23], and AsterDB[24], which enable searches for chemical structures by plant species names and other taxonomic information. Nevertheless, some data are still lacking for the purpose of exact dereplication. Information such as exact mass and geographic data can be very important for this type of study.[25][26][27]
It is not enough simply to focus on the information contained in a database. A clean and user-friendly interface, fast search, and consistency between currently available operating systems (Microsoft Windows, Mac, and Linux) can be just as important. For this purpose, the SistematX software was developed to provide the abovementioned information for chemosystematics studies, dereplication, and botanical correlations.
Results and discussion
Utility and discussion
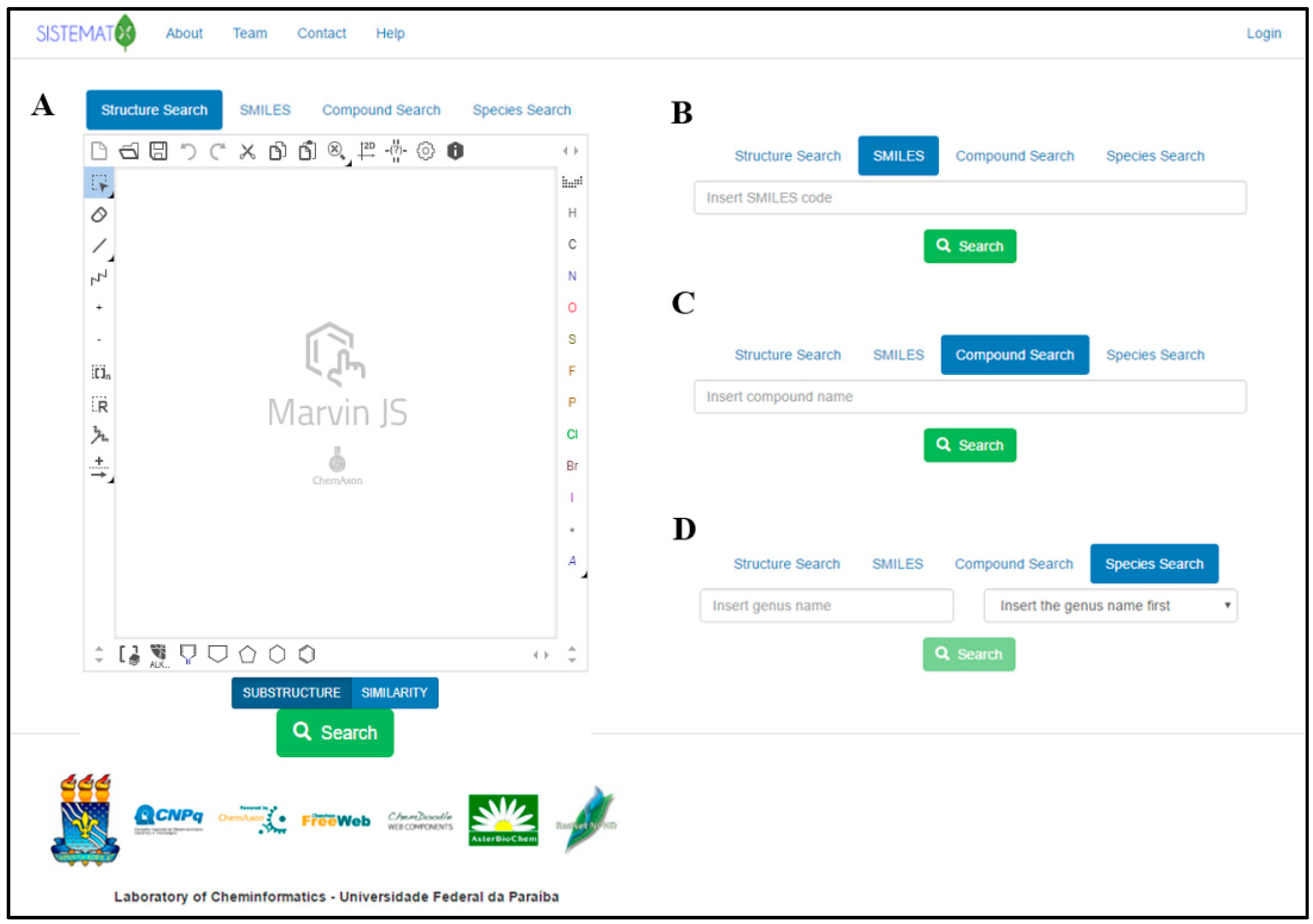
The SistematX homepage is shown in Figure 1A. After the user enters the website (http://sistematx.ufpb.br), the “Structure search” option is seen with the MarvinJS API (Application Programming Interface) at the top of the screen. Another three search options can be exhibited in the interface. The initial screen of the system also shows the SMILES (Simplified Molecular-Input Line-Entry System) code (Figure 1B), compound name (Figure 1C) and plant species search modes (Figure 1D).
|
In the first option, the user can perform the search using the drawn full structure or molecular skeleton, fragments, or substructures, which is important in cases when the user only knows a structural characteristic of the structure such as functional groups or when studies require structural similarity, structural groups, or families of compounds. It is possible to use the similarity search option that is currently available in SistematX. The search results page shows all compounds that correspond to the value above the cut off provided by the user in the decreasing order of similarity, showing the similarity values on the top. A substructure and similarity search is performed using a hashed fingerprint. Special molecular features are present in the query (e.g., stereochemistry, charge), only those targets match that also contain the feature. However, if a feature is missing from the query, it is not required to be missing.
In addition, it is possible to search by SMILES code, a chemical notation system capable of representing even the most complex organic compounds using a simple grammar that is very well known to organic chemistry researchers; for this reason we add this option separately from the structure search using the MarvinJS API, being friendly for one just to copy and paste the SMILES code (Figure 1B); by common (usual) name or IUPAC (International Union of Pure and Applied Chemistry) name (or part of one of these); and by species, although in this option, it is necessary to first insert the name of the genus (which presents an autocompletion option). After being selected, the system presents all species available for the user to select for the search.
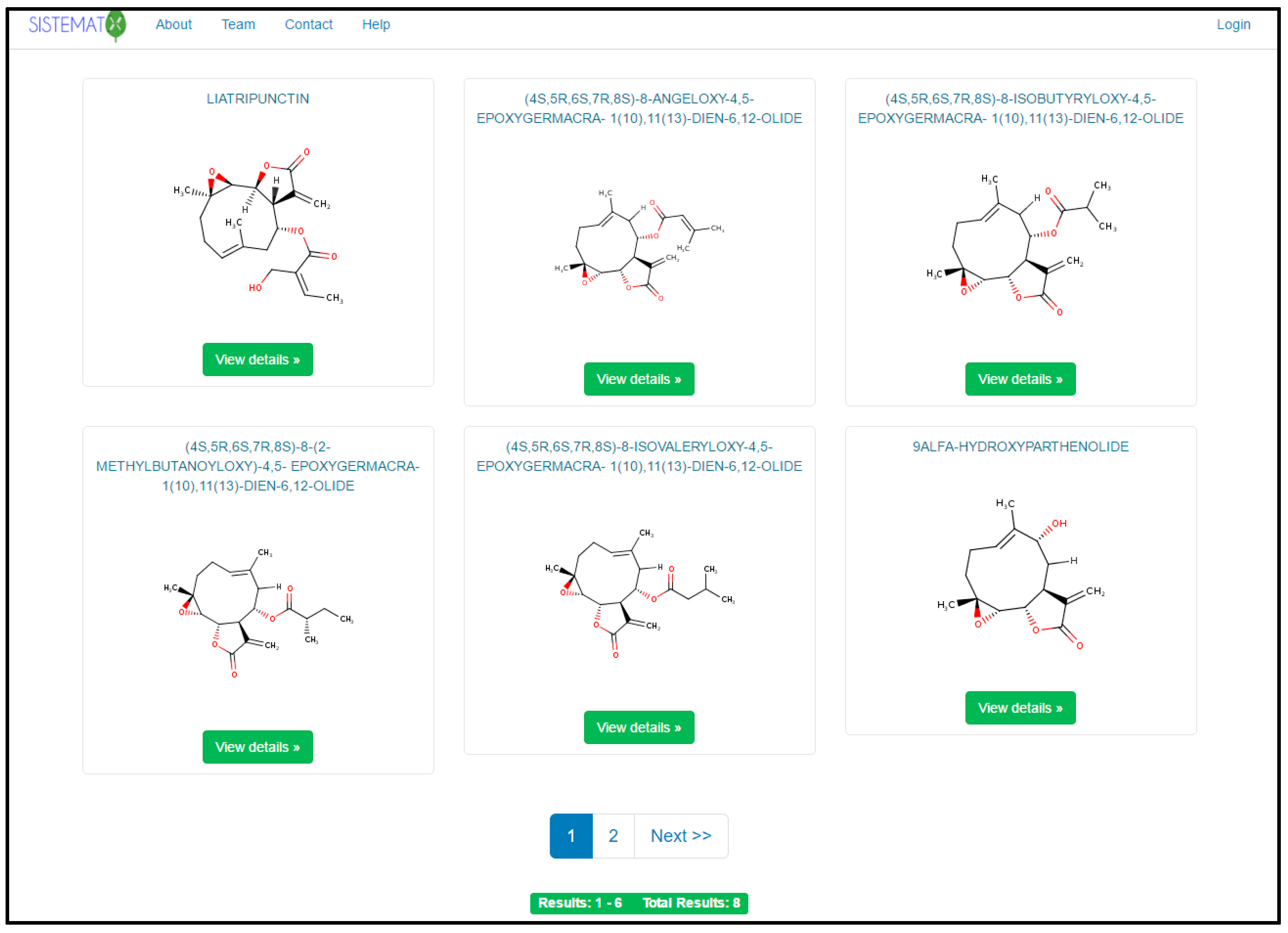
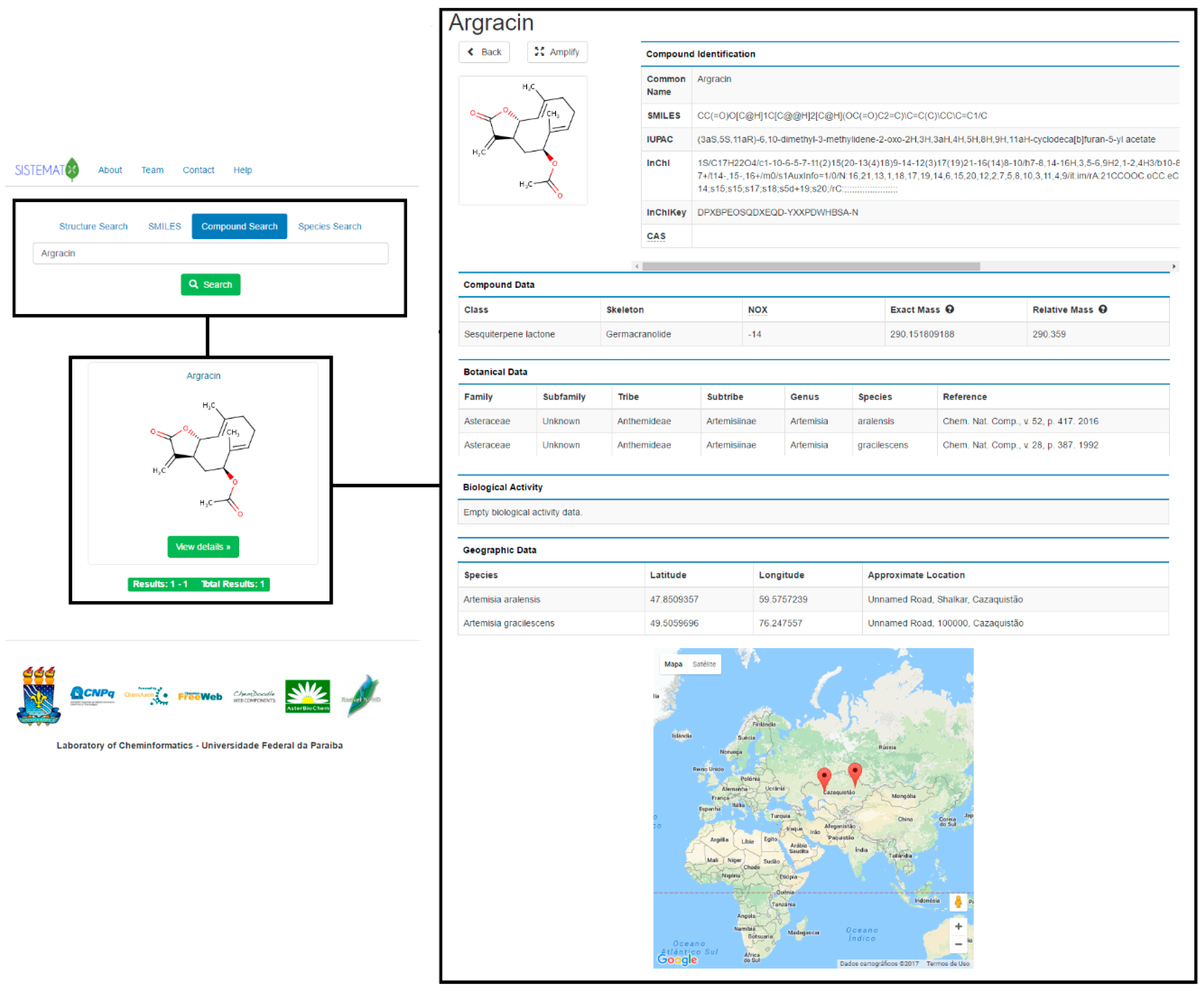
When performing a search, the mechanism generates a search results page (six results per page), using common names; if the compound does not have one, it shows the IUPAC name (Figure 2). The user can set the number of structure results per page. When a result is selected, the user has access to the data for that molecule, which are classified into six different groups (Figure 3).
|
|
The first group of results that appears is related to the structural representation of the searched molecule. The 2D structure is observed in the interface; on top, this appears as the option to amplify. After this is clicked, the system displays the visualization of the molecule in 2D and 3D (ChemDoodle, iChemLabs, Piscataway, NJ, USA) and an additional option for saving the 2D or 3D structure in an MDL (Molecular Design Limited, San Ramon, CA, USA) Molfile. The second type of result exhibited by the systems associated with compound identification, such as common name, SMILES code, IUPAC name, InChI (IUPAC International Chemical Identifier, Research Triangle Park, NC, USA) code, InChIKey code and CAS (Chemical Abstracts Service, Columbus, OH, USA) number. Except for the common name, which is optional and registered by the administrator, all parameters are provided by the JChem API.
Compound data results include important characteristics for natural products chemistry. The class of secondary metabolite of the searched molecule and its skeleton provide information about its biosynthetic pathway and assists in chemosystematics and chemotaxonomic studies. Oxidation number (NOX), which is calculated based on the Hendrickson rules[28], has been fundamental in chemotaxonomy since Gottlieb related the oxidation grade of molecules to species evolution.[29] Molecular mass is calculated using the most abundant isotope of each element (exact mass) and the average atomic mass of each element (relative mass); these data are important for users working on purification processes and for structural elucidation of molecules, due to the mass information, which is essential for determining the purity of secondary metabolites.
In the botanical data field, the user can find specific information such as the taxonomic rank (from family to species) of the plant from which the compound was isolated, the searched molecule, and the bibliographic reference, which includes journal name, volume, page, and year. Because many different species can biosynthesize the same molecule, there is one register per species. Meanwhile, the biological data exhibit results obtained in studies related to the biological activity of the searched molecule, the type of activity, system, units, activity value and bibliographic reference are available in this section.
Plant species have revealed clear genetic signals for local adaptation.[30] One species can synthesize a secondary metabolite depending on its location, and there are observed variations in compound concentrations at different sites. Because geographical data is an important parameter in natural products research, SistematX shows geographical coordinates (latitude and longitude) for a searched molecule and an approximate location of the species from which this metabolite was isolated. Using the Google Maps API, the user can observe the species location on the world map.
Data management
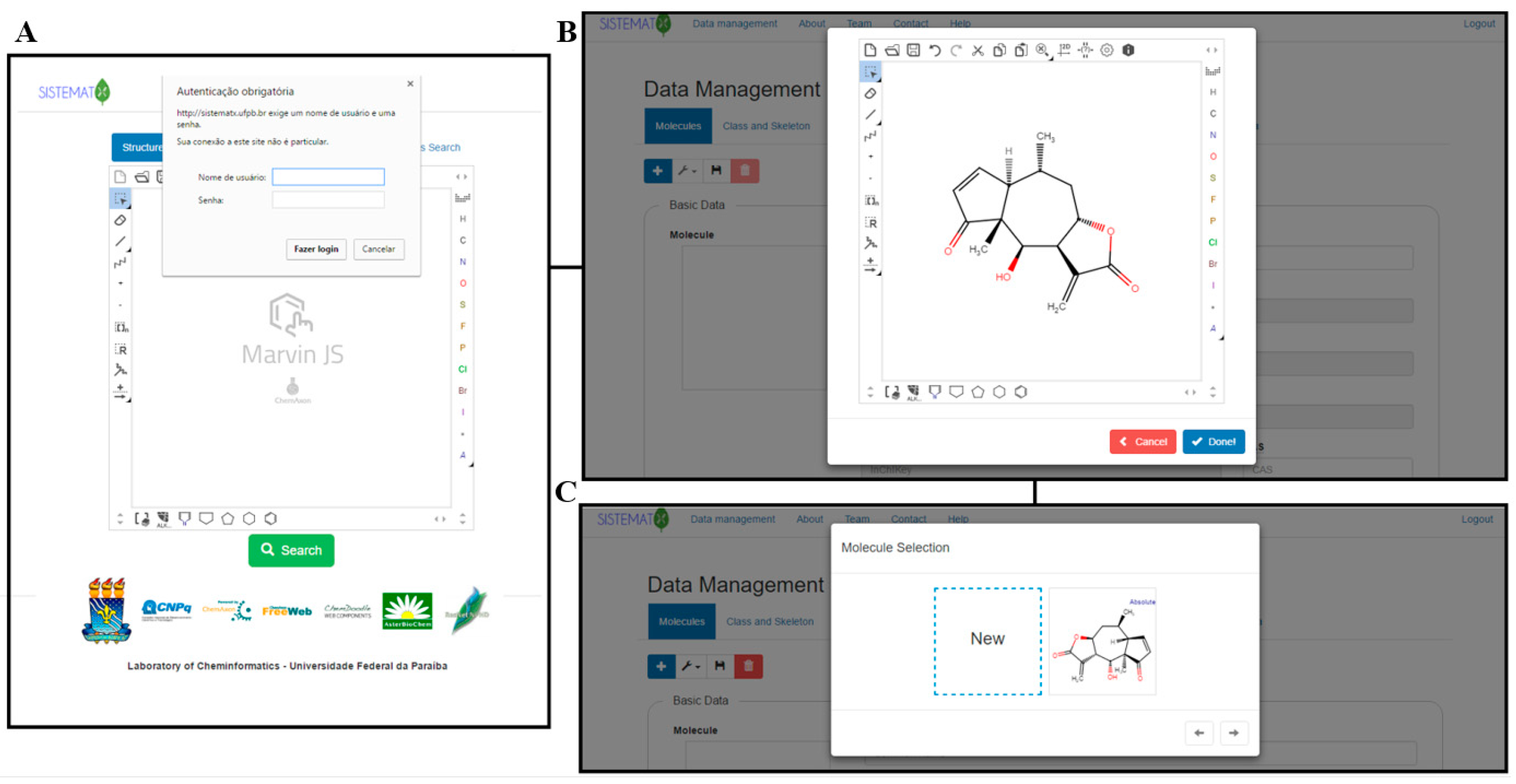
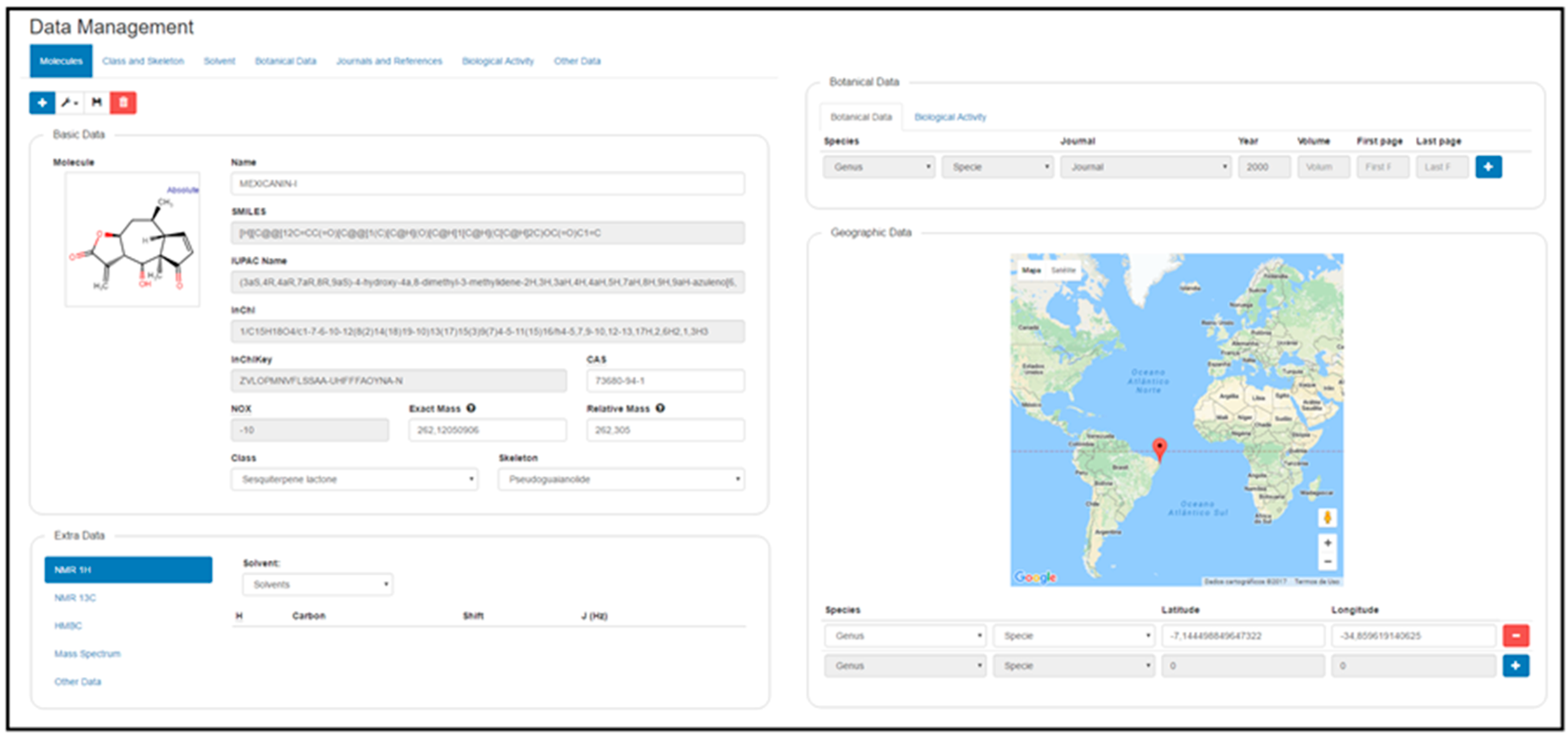
On the SistematX homepage, the user can also log into the data management area using login name and password (Figure 4A) and from there access the administration pages to edit or register new molecules. Once the corresponding information has been accepted, the data management interface appears.
|
The first requirement to register a new molecule is to insert the structure in the MarvinJS API (Figure 4B). Several methods can be used to accomplish this step: drawing, copying SMILES code, or importing the molecule in a compatible format (e.g., sdf, cdx, mol, mol2). The molecule selection option then appears (Figure 4C), and if the molecule is new to the system, it appears with the option "New." If this option is selected, a blank register page with four subdivisions is shown: Basic Data, Extra Data, Botanical Data, and Geographical Data. However, if the molecule already exists in the system, another box with the drawn molecule appears, and after choosing this box, the register page for the structure containing all previously registered information appears (Figure 5).
|
Immediately after the New option is selected, it appears in the register page, including some basic data generated by the MarvinJS API: SMILES, IUPAC name, InChI, InChIKey, NOX, exact mass, and relative mass. The class and skeleton of metabolites must be chosen to register a new structure in the system; this is a sequential process, and thus, the class must first be selected to make the skeleton structure option available. Classes and skeletons not already registered in the system can be registered via the Class and Skeleton tab on the top of the screen. Common name and CAS are optional registration options.
The Extra Data subdivision allows insertion of all structural spectroscopic information, such as 1H- and 13C-NMR (Nuclear Magnetic Resonance) and mass spectra. In addition, it is possible to find 2D NMR information through the HMBC (Heteronuclear Multiple Bond Correlation) technique to establish the relationship between 13C- and 1H-shifts. In the NMR data, the administrator must first select the deuterated solvent used in the spectroscopic studies. If this information does not exist in the options, it can be registered by selecting the Solvent tab on the top of the screen. After the structure appears with an atomic numeration assigned by the MarvinJS API (identified as Atom), it is always necessary to verify these numbers for the biogenetic numeration (identified as “Biogenetic”) and finally to add the chemical shift value for each atom. For 1H-NMR, it is also possible to register H–H coupling constants (J in Hz). Mass spectrum information, molecular mass, and intensity of fragments can also be being registered.
To create a new registry for botanical data of a certain species, the following information is required: journal, year, volume, first page, and last page information. Journal, genus, and species are drop-down lists, and these last two must be filled in this order. If any information relevant to these three fields does not exist, it can be inserted by clicking in "Journal" and "References" and writing the journal name (autocomplete tab) or by clicking in "Botanical Data," where taxonomic data are registered. Biological activity data appear as activity, system, system type, value, journal, year, and pages. Drop-down lists can be filled with previously registered data or by entering new data in the Biological Activity tab.
Finally, the administrator can register geographical data using the Google maps API. For any structure, it is possible to register latitude and longitude of the corresponding species studied. The genus and species boxes are filled in the same manner described above. Longitude and latitude can be inserted in two ways, first by writing the coordinates in the spaces; once registered, they appear on the world map as a red indicator showing the location. Another method is to select the place by clicking on the red pin on the map; when the pin is released, the values of longitude and latitude appear in the respective boxes.
Currently, our database has more than 1300 sesquiterpene lactones and 850 flavonoids and chalcones, with more than 4000 botanical occurrences of the Asteraceae family; approximately 500 alkaloids, which represents more than 750 botanical occurrences of the Apocynaceae family; and several terpenes and alkaloids of Annonaceae, Apocynaceae, and Asteraceae that correspond to more than 800 botanical occurrences.
Material and methods
Implementation
SistematX was developed in the Java programming language (version 8 or higher), using JSP (JavaServer Pages) technology version 2.1 or higher and MySQL database version 5.5.46-0 for Linux[31] to maintain the system data. An initial version of SistematX web was published in the proceedings of MOL2NET in 2015 to demonstrate its functionalities to the academic community and, with feedback, to improve old functionalities and add new tools.[32] In the current version, including automatically relative mass, exact mass, CAS number, and InChI (in the previous version only the InChIKey was available) for each compound. The CAS number if not generated automatically but can be added manually, and geographical localization of species where the compound was isolated is now available. It is also possible to perform a structural search by similarity index.
The system uses JSP to create pages with specific information for each molecule and dynamic page changes by clicking on certain buttons. Intermediary pages are used to recover information from the database and insert it in the JSP. The system creates a DAO (data access object) to organize the data on intermediary pages, working like a bridge from the DAO to JSP. For each database table needed in a request, a DAO is created specifically for that table, containing all attributes from the database table.
Bootstrap version 3.3.5[33] — a graphical interface web framework that uses HTML (HyperText Markup Language), CSS (Cascading Style Sheets) and JavaScript — is employed for a better appearance on HTML pages, adapting some functions and styles to aid in the website design. The system also uses jQuery framework version 2.1.4[34], a powerful JavaScript tool to manipulate HTML DOM (document object model) events. Another framework, jQuery AutoComplete version 1.2.18[35], is used to generate autocomplete inputs.
Several APIs are used in the SistematX implementation (Table 1). MarvinJS version 15.7.20, from ChemAxon[36], is the drawing API that is integrated with ChemAxon JChem WebService[37], an external online service that transforms the drawn structure into SMILES code, after which a JChem API function turns it into a binary fingerprint. This fingerprint is used to search for molecules — using substructure or similarity — in the database via their structure. The molecule converted to the fingerprint is used as a fragment in the search, comparing it to the database molecule’s fingerprints to determine if it exists inside as a fragment. This API utilizes HTML, CSS, and JavaScript to perform the transformations. We use the standardizer API of Chemaxon JChem Web Services to not only standardize nitro groups but also aromatize the structure search results as correctly as possible, making a query and the database with similar representation.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
MarvinJS is used to create a 3D structure from a 2D molecule, and ChemDoodle Web Components[38] shows this view as JavaScript in the browser. This API is able to generate several 2D and 3D molecule graphical views using pure JavaScript.
The ChemAxon API allows the visualization of compound characteristics. SistematX displays general nomenclature information such as the common name, SMILES code, IUPAC name, InChI, InChIKey, CAS registry number and properties such as oxidation number (NOX), exact mass, and relative mass.
In addition, Google Maps, from Google Inc.[39], an API used to prepare maps and locations, is used in the system to show the registered metabolite location on the world map. The API draws the map and receives locations from the database, which are two variables representing the latitude and longitude. A registered molecule may have multiple locations and a species linked to it. Using a JavaScript function, it graphically sets the locations on the map. When registering by clicking on the map, it sets a marker at the mouse location and adds a line to the coordinates list below the map for each marker on the map, allowing it automatically to change the position when the value in the latitude or longitude boxes is changed. The coordinates are also transformed into an approximate address, using reverse geocode, a function from the Google Maps API.
Conclusions
In this article, we introduce a web interface for managing a secondary metabolite database, which is multiplatform and able to be consulted via the internet and managed from any accredited computer. The interface provides a wealth of useful information for the scientific community about natural products, highlighting the location of species from which the compounds were isolated. Several new functionalities will be added continuously, such as new calculated molecular descriptors, tools to aid structural elucidation using experimental and calculated NMR data, and support for downloading several structures in one file using a batch transfer data option.
Sistemat X is freely accessible on the homepage http://sistematx.ufpb.br.
Acknowledgments
We would like to thank the Student Agreement Program of Graduate—PEC-PG of CNPq, Brazil and the Brazilian National Council for Scientific and Technological Development (CNPq)—Award Number 461093/2014-6.
Author contributions
S.Y.K.d.O.S., R.P.O.C. and M.T.S. carried out the design, programing and drafted the manuscript. C.H.-A., T.B.O., F.B.D.C., L.S. and R.P.R. carried out the design, tests and drafted the manuscript. C.H.-A., L.S., F.B.D.C. and M.T.S. revised the manuscript critically.
Conflicts of interest
The authors declare no conflict of interest.
References
- ↑ Blunt, J.W.; Munro, M.H.G. (2014). "22. Is There an Ideal Database for Natural Products Research?". In Osbourn, A.; Goss, R.J.; Carter, G.T.. Natural Products: Discourse, Diversity, and Design. John Wiley & Sons, Inc. pp. 413–431. doi:10.1002/9781118794623.ch22. ISBN 9781118794623.
- ↑ 2.0 2.1 Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. (2015). "The re-emergence of natural products for drug discovery in the genomics era". Nature Reviews Drug Discovery 14 (2): 111–29. doi:10.1038/nrd4510. PMID 25614221.
- ↑ Corley, D.G.; Durley, R.C. (1994). "Strategies for Database Dereplication of Natural Products". Journal of Natural Products 57 (11): 1484–1490. doi:10.1021/np50113a002.
- ↑ Oliveira, T.; Chagas-Paula, D.; Rosa, A. et al. (2013). "Temporal characteristics of a natural products in-house database". Planta Medica 79: 1113–1114. doi:10.1055/s-0033-1351852.
- ↑ Pence, H.E.; Williams, A. (2010). "ChemSpider: An Online Chemical Information Resource". Journal of Chemical Education 87 (11): 1123–1124. doi:10.1021/ed100697w.
- ↑ Bolton, E.E.; Wang, Y.; Thiessen, P.A.; Bryant, S.H. (2008). "Chapter 12 - PubChem: Integrated Platform of Small Molecules and Biological Activities". In Wheeler, R.A.; Spellmeyer, D.C.. Annual Reports in Computational Chemistry. 4. Elsevier Ltd. pp. 217-241. doi:10.1016/S1574-1400(08)00012-1. ISBN 9780444532503.
- ↑ Degtyarenko, K.; de Matos, P.; Ennis, M. et al. (2008). "ChEBI: A database and ontology for chemical entities of biological interest". Nucleic Acids Research 36 (DB1): D344-50. doi:10.1093/nar/gkm791. PMC PMC2238832. PMID 17932057. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2238832.
- ↑ Irwin, J.J.; Shoichet, B.K. (2005). "ZINC: A free database of commercially available compounds for virtual screening". Journal of Chemical Information and Modeling 45 (1): 177–82. doi:10.1021/ci049714+. PMC PMC1360656. PMID 15667143. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1360656.
- ↑ 9.0 9.1 Williams, R.B.; O'Neil-Johnson, M.; Williams, A.J. et al. (2015). "Dereplication of natural products using minimal NMR data inputs". Organic & Biomolecular Chemistry 13 (39): 9957–62. doi:10.1039/c5ob01713k. PMID 26381222.
- ↑ Eugster, P.J.; Boccard, J.; Debrus, B. et al. (2014). "Retention time prediction for dereplication of natural products (CxHyOz) in LC-MS metabolite profiling". Phytochemistry 108: 196–207. doi:10.1016/j.phytochem.2014.10.005. PMID 25457501.
- ↑ "Dictionary of Natural Products". CRC Press. http://dnp.chemnetbase.com/. Retrieved 04 March 2017.
- ↑ Graham, J.G.; Farnsworth, N.R. (2010). "The NAPRALERT database as an aid for discovery of novel bioactive compounds". Comprehensive Natural Products II: Chemistry and Biology. 3. Elsevier Ltd. pp. 81–94. ISBN 9780080453828.
- ↑ Dabb, S.; Blunt, J.; Munro, M. (2014). "MarinLit: Database and essential tools for the marine natural products community". Proceedings of the 248th National Meeting of the American-Chemical-Society (ACS) 248.
- ↑ Banerjee, P.; Erehman, J.; Gohlke, B.O. et al. (2015). "Super Natural II:A database of natural products". Nucleic Acids Research 43 (DB1): D935–9. doi:10.1093/nar/gku886. PMC PMC4384003. PMID 25300487. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4384003.
- ↑ Drewry, D.H.; Macarron, R. (2010). "Enhancements of screening collections to address areas of unmet medical need: An industry perspective". Current Opinions in Chemical Biology 14 (3): 289–98. doi:10.1016/j.cbpa.2010.03.024. PMID 20413343.
- ↑ Valli, M.; dos Santos, R.N.; Figueira, L.D. et al. (2013). "Development of a natural products database from the biodiversity of Brazil". Journal of Natural Products 76 (3): 439-44. doi:10.1021/np3006875. PMID 23330984.
- ↑ Hatherley, R.; Brown, D.K.; Musyoka, T.M. et al. (2015). "SANCDB: A South African natural compound database". Journal of Cheminformatics 7: 29. doi:10.1186/s13321-015-0080-8. PMC PMC4471313. PMID 26097510. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4471313.
- ↑ Kim, S.K.; Nam, S.; Jang, H. et al. (2015). "TM-MC: A database of medicinal materials and chemical compounds in Northeast Asian traditional medicine". BMC Complementary and Alternative Medicine 15: 218. doi:10.1186/s12906-015-0758-5. PMC PMC4495939. PMID 26156871. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4495939.
- ↑ Chen, C.Y. (2011). "TCM Database@Taiwan: The world's largest traditional Chinese medicine database for drug screening in silico". PLoS One 6 (1): e15939. doi:10.1371/journal.pone.0015939. PMC PMC3017089. PMID 21253603. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3017089.
- ↑ Ntie-Kang, F.; Telukunta, K.K.; Döring, K. et al. (2017). "NANPDB: A Resource for Natural Products from Northern African Sources". Journal of Natural Products 80 (7): 2067-2076. doi:10.1021/acs.jnatprod.7b00283. PMID 28641017.
- ↑ Xue, R.; Fang, Z.; Zhang, M. et al. (2013). "TCMID: Traditional Chinese Medicine integrative database for herb molecular mechanism analysis". Nucleic Acids Research 41 (DB1): D1089-95. doi:10.1093/nar/gks1100. PMC PMC3531123. PMID 23203875. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3531123.
- ↑ Afendi, F.M.; Okada, T.; Yamazaki, M. et al. (2012). "KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research". Plsnt & Cell Physiology 53 (2): e1. doi:10.1093/pcp/pcr165. PMID 22123792.
- ↑ Tung, C.W.; Lin, Y.C.; Chang, H.S. et al. (2014). "TIPdb-3D: The three-dimensional structure database of phytochemicals from Taiwan indigenous plants". Database 2014: bau055. doi:10.1093/database/bau055. PMC PMC4057645. PMID 24930145. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4057645.
- ↑ "AsterDB". AsterBioChem. http://www.asterbiochem.org/asterdb. Retrieved 04 March 2017.
- ↑ Sampaio, B.L.; Edrada-Ebel, R.; Da Costa, F.B. (2016). "Effect of the environment on the secondary metabolic profile of Tithonia diversifolia: A model for environmental metabolomics of plants". Scientific Reports 6: 29265. doi:10.1038/srep29265. PMC PMC4935878. PMID 27383265. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4935878.
- ↑ Schmidt, T.J.; Rzeppa, S.; Kaiser, M.; Brun, R. (2012). "Larrea tridentata—Absolute configuration of its epoxylignans and investigations on its antiprotozoal activity". Phytochemistry Letters 5 (3): 632-638. doi:10.1016/j.phytol.2012.06.011.
- ↑ Gaquerel, E.; Kuhl, C.; Neumann, S. (2013). "Computational annotation of plant metabolomics profiles via a novel network-assisted approach". Metabolomics 9 (4): 904–918. doi:10.1007/s11306-013-0504-2.
- ↑ Hendrickson, J.B.; Cram, D.J.; Hammond, G.S. (1970). Organic Chemistry. McGraw-Hill. ISBN 070281505.
- ↑ Gottlieb, O. (1989). "The role of oxygen in phytochemical evolution towards diversity". Phytochemistry 28: 2545–2558.
- ↑ Züst, T.; Heichinger, C.; Grossniklaus, U. et al. (2012). "Natural enemies drive geographic variation in plant defenses". Science 338 (6103): 116–9. doi:10.1126/science.1226397. PMID 23042895.
- ↑ "Download MySQL Community Server". Oracle Corporation. https://dev.mysql.com/downloads/mysql/. Retrieved 31 January 2017.
- ↑ Scotti, M.T.; Da Silva Junior, R.O.; De Oliveira Santos, S.Y.K. et al. (2015). [https://sciforum.net/conference/MOL2NET-1/paper/3348/download/pdf "SISTEMAT X - A Web Tool to Manage Databases of Secondary Metabolites"]. Proceedings of MOL2NET, International Conference on Multidisciplinary Sciences 1 (Section F): 1–6. https://sciforum.net/conference/MOL2NET-1/paper/3348/download/pdf.
- ↑ "Bootstrap". Bootstrap Development Team. http://getbootstrap.com/. Retrieved 10 January 2017.
- ↑ "jQuery - Write less, do more". The jQuery Foundation. http://jquery.com/. Retrieved 17 January 2017.
- ↑ "jQuery - User interface". The jQuery Foundation. http://jqueryui.com/autocomplete/. Retrieved 20 January 2017.
- ↑ "Marvin JS". ChemAxon Ltd. https://chemaxon.com/products/marvin-js. Retrieved 24 January 2017.
- ↑ "JChem Engines". ChemAxon Ltd. https://chemaxon.com/products/jchem-engines. Retrieved 24 January 2017.
- ↑ "ChemDoodle Web Components". iChemLabs, LLC. https://web.chemdoodle.com/. Retrieved 24 January 2017.
- ↑ "Google Maps APIs". Google Inc. https://developers.google.com/maps/. Retrieved 13 January 2017.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.