Journal:The systems biology format converter
| Full article title | The systems biology format converter |
|---|---|
| Journal | BMC Bioinformatics |
| Author(s) |
Rodriguez, N.; Pettit, J.-B.; Pezze, P.D.; Li, L.; Henry, A.; van Iersel, M.P.; Jalowicki, G.; Kutmon, M.; Natarajan, K.N.; Tolnay, D.; Stefan, M.I.; Evelo, C.T.; Le Novère, N. |
| Author affiliation(s) |
EMBL European Bioinformatics Institute, The Babraham Institute, Maastricht University, California Institute of Technology |
| Primary contact | E-mail: lenov@babraham.ac.uk |
| Year published | 2016 |
| Volume and issue | 17 |
| Page(s) | 154 |
| DOI | 10.1186/s12859-016-1000-2 |
| ISSN | 1471-2105 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-016-1000-2 |
| Download | http://bmcbioinformatics.biomedcentral.com/track/pdf/10.1186/s12859-016-1000-2 (PDF) |
Abstract
Background
Interoperability between formats is a recurring problem in systems biology research. Many tools have been developed to convert computational models from one format to another. However, they have been developed independently, resulting in redundancy of efforts and lack of synergy.
Results
Here we present the System Biology Format Converter (SBFC), which provide a generic framework to potentially convert any format into another. The framework currently includes several converters translating between the following formats: SBML, BioPAX, SBGN-ML, Matlab, Octave, XPP, GPML, Dot, MDL and APM. This software is written in Java and can be used as a standalone executable or web service.
Conclusions
The SBFC framework is an evolving software project. Existing converters can be used and improved, and new converters can be easily added, making SBFC useful to both modellers and developers. The source code and documentation of the framework are freely available from the project web site.
Keywords
Converter, Format, Systems biology, SBML
Background
Computational representations of pathways and models lie at the core of systems biology research.[1] Formats have been designed to encode these complex knowledge representations, either as community standards or as formats specific to a software tool.[2] Different formats are preferentially used to address specific problems or use different approaches, thus limiting interoperability. However, one often needs to use several tools and approaches to answer a biological question, or to reuse existing pathways and models in different contexts. Many format converters have been written over the years. Often, several converters between the same formats are developed independently by different groups. This results in a duplication of efforts and waste of time, energy and money. The different converters may be inconsistent, leading to different results. In addition, being developed by one person or one team, those software tools tend to go unmaintained while the formats they are covering keep evolving. Finally, some of these converters are embedded in larger pieces of software, which hinders their use.
To overcome these challenges, the Systems Biology Format Converter (SBFC) software provides an open-source modular and extensible framework to potentially support conversion between any two formats using a single executable or web service.
Implementation
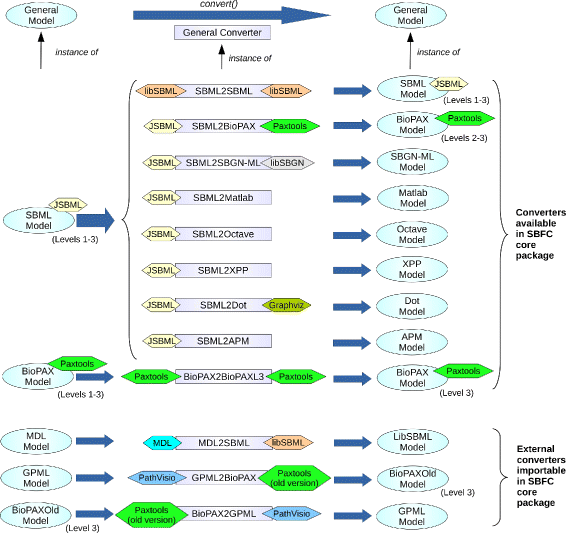
SBFC was built to support rapid implementation and integration of new converters. Therefore, it was designed with a high degree of modularity. At the core of the software are the GeneralModel interface and the GeneralConverter abstract class. The former is used for data exchange and describes the operations that every input or output format object must implement to be processed by SBFC. The latter represents the generic algorithm for converting one format into another. An overview of the SBFC framework is provided on Fig. 1.
|
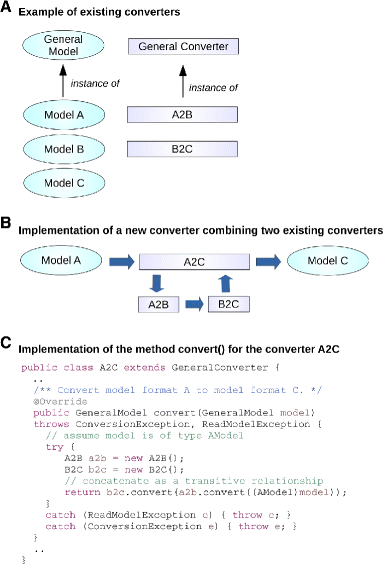
To add a new format, a developer must simply implement the GeneralModel interface, which provides some methods to read and write the format to file or string. Adding a new converter requires extending the GeneralConverter class and implementing the GeneralModel convert(GeneralModel model) method, where the model parameter is the input format that needs to be converted and the returned GeneralModel object is the new converted format. For instance a converter A2B translating from a file formatted as model A to a file formatted as model B requires the definition of two classes ModelA and ModelB implementing the GeneralModel interface. The class converter A2B must extend the abstract class GeneralConverter and implement the method GeneralModel convert(GeneralModel model). This method will receive an input object named model, whose dynamic type is ModelA. The object returned by this method will have dynamic type ModelB.
Because all SBFC format classes are implementations of the GeneralModel interface, it is possible to create new converters re-using existing converters by simply invoking the generic convert() method for each existing converter (Fig. 2). The convert() method in the new converter A2C is implemented by calling the convert() methods in the converters A2B and B2C, respectively (source code for all classes is available in the SBFC manual).
|
SBFC is developed using the Java programming language. However, if an existing converter is developed in a programming language other than Java, it is still possible to create a new SBFC converter that will invoke the existing converter using the Java Runtime exec() method. This approach can be used for invoking any external program or command without having to re-write the full converter. Once the converter is integrated into the framework, it can be used and combined effortlessly with other converters (source code available in the SBFC manual). A potential disadvantage of this approach is the loss of interoperability when using operating system-dependent code. The advantage is that the specific SBFC converters directly rely on the development of the original external converters reducing code duplication.
Each format is identified by an identifiers.org URI[3] or an internet media type. If none of them exists, the developers of the format and converter classes must agree on an identifier (URI) for this format. SBFC allows multiple classes implementing the GeneralModel interface for a given format, using different tools to read and write models. All classes should return the same value for the getURI() method. For instance, the implementation of a converter for the Systems Biology Markup Language[4] may rely on JSBML[5], libSBML[6], or a DOM document structure.[7] This design can be advantageous when 1) a given library does not read a version of a format properly; 2) a converter was written with an old or newer version of a library that has a different API; or 3) high performance is required (e.g. improving the implementation for file processing). At the beginning of a conversion, the converter checks that the value returned by the getURI() method of the input GeneralModel is a URI of a format it does support. If the converter recognises the format URI, the generic write methods (modelToString() or modelToFile(String fileName)) are used in order to retrieve the file content.
Results
Available formats and converters
The SBFC project already implemented support for several formats and developed several converters. The following format classes are provided:
- APMModel for the APMonitor Modelling Language (APM). APMonitor is an optimization software for mixed-integer and differential algebraic equations[8];
- BioPAXModel for BioPax, format to exchange descriptions of biomolecular pathways, including reaction and interaction networks[9];
- DotModel for the Dot format, that encodes graph descriptions used by the open source graph visualisation software GraphViz[10] to generate multiple image formats (e.g. PNG, JPEG, etc);
- GPMLModel for the format used by the pathway drawing and analysis tool PathVisio[11] and the pathway database WikiPathways[12];
- MDLModel for the format used by the single particle simulator MCell[13];
- OctaveModel for Octave and MatLab m-file formats, encoding mathematical models usable by the modeling environments GNU Octave[14] and MatLab;
- SBGNMLModel for SBGN-ML format, a format to encode graphical maps in the Systems Biology Graphical Notation[15];
- SBMLModel for SBML[4], a format encoding mathematical models;
- XPPModel for XPP format, encoding mathematical models usable by the numerical analysis software XPPAUT.[16]
This core set of model formats was based on the set of converters internally used by the BioModels project.[17] As SBML is central for this resource, the focus was on implementing converters from SBML to other formats. The following converters listed by class name are currently available: SBML2SBML, SBML2APM, SBML2BioPAX, SBML2SBGNML, SBML2Matlab, SBML2Octave, SBML2XPP, SBML2Dot. SBFC also supports conversions between SBML model annotations urn:miriam[18] and Identifiers.org URL[3] with the converters URN2URL and URL2URN. The Systems Biology community has been developing additional converters, including MDL2SBML, GPML2BioPAX, BioPAX2BioPAXL3 (converting from BioPAX Level 1 and 2 to BioPAX Level 3).
Running SBFC as a standalone application
SBFC can be used as a standalone application and executed via a basic graphical user interface or the command line. The general command to convert one model into another on the command line is provided by the Converter class within the package org.sbfc.converter. The syntax for this generic command is:
java org.sbfc.converter.Converter [InputModelClass] [ConverterClass] [ModelFile]
For instance, a model in SBML format can be converted to Octave format using the command:
java org.sbfc.converter.Converter SBMLModel SBML2Octave model.xml
Bash and bat scripts are provided for most converters. This removes the need to specify the Java classpath, the input class, and converter classes. The previous conversion becomes:
./sbml2octave.sh my-sbml-model.xml
Users can specify a folder name as parameter, instead of a model file. In this case, the script will try to convert all xml files found in the folder.
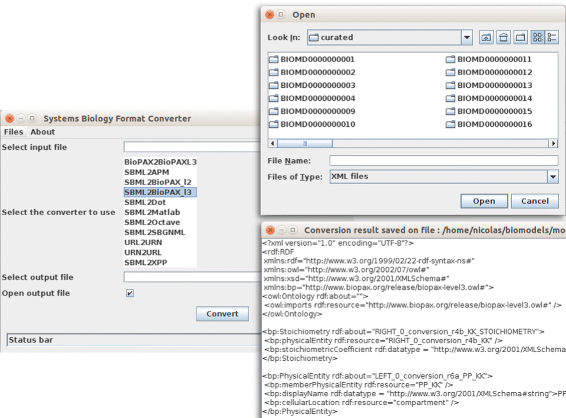
If the number of files to be converted is limited, a simple standalone GUI can also be used (Fig. 3). The general syntax is:
java -jar sbfc-x.y.z-standalone.jar
|
On most systems, a double click on the Jar is sufficient but some scripts are also provided to help users to launch the GUI:
./sbfConverterGUI.sh for Linux or sbfConverterGUI.bat on Windows
This GUI can be embedded in any 3rd-party Java software with the command:
ConverterGUI.getConverterGuiInstance().setVisible(true);
Running SBFC online
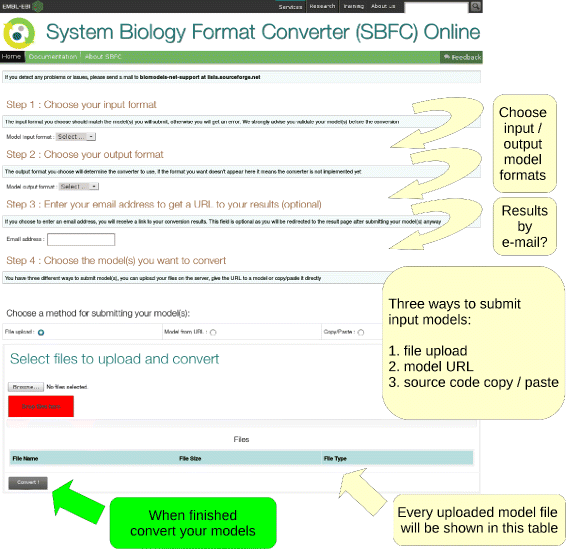
To provide easy access to SBFC, a web application has been deployed at the European Bioinformatics Institute (EMBL-EBI).[19] The web page is used in four successive steps as shown in Fig. 4. The user must first specify the input and output model formats. Once the input format is selected, the list of possible output formats is updated depending of the converters available on the system. An e-mail address can be optionally provided to receive a URL link to these results. Finally, the documents to convert can be selected. This can be done via three methods: file upload, model URL or copy/paste. Once a conversion is launched, the user is redirected to a result page. All the jobs launched during a browser session will be displayed. The results can be downloaded for 72 h. It is worth noting that for privacy purposes neither the original nor the converted models are kept on the servers for a period longer than 72 h.
|
The SBFC Online application can be downloaded from the Sourceforge website and installed locally as described in the SBFC developer manual.
Running SBFC through Web services
For automatic and programmatic access, SBFC provides REST like webservices using the HTTP POST method. For java users, a helper class SBFCWebServiceClient provides an API that simplifies the use of the Web Services. Depending on user requirements, two types of methods can be selected for submitting conversion jobs. The first type performs blocking calls. The methods submitAndGetResultFromFile, submitAndGetResultFromURL and submitAndGetResultFromString start a conversion job and wait until the model is converted and returned from the web server. For large models, the conversion process can last several minutes, particularly if the cluster load is high. The second type of methods use asynchronous, nonblocking calls. The methods submitJobFromFile, submitJobFromURL and submitJobFromString start a conversion job and immediately return a ConversionId object that contain some metadata about the job. The status of the job can be checked later with the getJobStatus method. When the returned status is ‘done’, the user can use the method getConvertionResult to retrieve the output file. An example of java code that launches several conversions through the SBFC Web Services is provided in the additional file UsageExample.java [see Additional file 1].
Discussion
Novel formats are created on a regular basis in systems biology, due to the development of new software tools for building and analysing biological pathways, networks and models. While these developments are necessary for the progress of the domain, interoperability is a crucial challenge and conversion tools play a central role. In order to maintain feature rich and up to date code, as well as to limit redundant efforts, community efforts are need. SBFC is an attempt in that direction.
SBFC was implemented in Java in order to be fully portable on all operating systems provided with a Java virtual machine. This facilitates software adoption by users as these do not have to install platform-specific versions, and by developers who can easily integrate SBFC in their existing Java software if needed. The provided web services also allow software not implemented in Java to use SBFC easily.
SBFC architecture is sufficiently generic to permit rapid implementations and additions of new formats and converters. With this solid but flexible design, SBFC aims to make existing efforts to develop systems biology converters converge into a single community activity.
A roadmap including new features and converters to be developed in the near future is available.[20] Contribution from the community is needed to integrate converters more quickly. An important feature which is still in progress is the complete adoption of the OSGi framework[21] within SBFC. This implementation would reduce problems related to library conflicts between different converters. The future SBFC OSGi plugins would also enable a complete integration of SBFC as Cytoscape 3[22] plugin.
Conclusions
SBFC is novel open-source software that provides a generic Java-based architecture for converting between Systems Biology model formats (but not limited to those). It helps computational biologists to process or visualise their models using different software tools, and software developers to implement format conversion. We hope that new converters will be contributed in the future.
Availability and requirements
Project name: Systems Biology Format Converter (SBFC)
Project home page: http://sbfc.sourceforge.net/
Operating system(s): Platform independent
Programming language: Java SE 6 or higher
Other requirements: None
License: GNU LGPL v2
Abbreviations
APM: APMonitor Modelling Language
BioPAX: Biological Pathway Exchange
GPML: Pathvisio Graphical Pathway Markup Language
MDL: MCell Modelling Description Language
SBFC: Systems Biology Format Converter
SBGN: Systems Biology Graphical Notation
SBML: Systems Biology Markup Language
URI: Uniform Resource Identifier
Declarations
Open access
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
Additional file
Additional file 1: SBFC Web Services usage example. The file UsageExample.java presents some code using several methods of the SBFCWebServiceClient class to perform some conversions, retrieve the results and save them to local files. (JAVA 5 kb)
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JBP, NLN, NR, MK and MPI conceived and designed the project; JPB, MK, MPI, NR and PDP implemented the software; in addition LL, AH, GK and KN contributed to some of the included converters. JPB, PDP, NLN and NR wrote the manuscript. All authors read and approved the final manuscript.
Funding
The work was funded by British BBSRC (BB/F010516/1, BBS/E/B/000C0419), EMBL-EBI and US National Institute of Health (GM070923C).
References
- ↑ Le Novère, N. (2015). "Quantitative and logic modelling of gene and molecular networks". Nature Reviews Genetics 16 (3): 146–158. doi:10.1038/nrg3885. PMC PMC4604653. PMID 25645874. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4604653.
- ↑ Hucka, M., Nickerson, D.P.; Bader, G.D. et al. (2015). "Promoting coordinated development of community-based information standards for modeling in biology: The COMBINE initiative". Frontiers in Bioengineering and Biotechnology 3: 19. doi:10.3389/fbioe.2015.00019. PMC PMC4338824. PMID 25759811. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4338824.
- ↑ 3.0 3.1 Juty, N.; Le Novère, N.; Laibe, C. (2012). "Identifiers.org and MIRIAM Registry: Community resources to provide persistent identification". Nucleic Acids Research 40 (D1): D580-D586. doi:10.1093/nar/gkr1097. PMC PMC3245029. PMID 22140103. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3245029.
- ↑ 4.0 4.1 Hucka, M.; Finney, A.; Sauro, H.M. et al. (2003). "The systems biology markup language (SBML): A medium for representation and exchange of biochemical network models". Bioinformatics 19 (4): 524-531. doi:10.1093/bioinformatics/btg015. PMID 12611808.
- ↑ Dräger, A.; Rodriguez, N.; Dumousseau, M. et al. (2011). "JSBML: A flexible Java library for working with SBML". Bioinformatics 27 (15): 2167-2168. doi:10.1093/bioinformatics/btr361. PMC PMC3137227. PMID 21697129. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3137227.
- ↑ Bornstein, B.J.; Keating, S.M.; Jouraku, A.; Hucka, M. (2008). "LibSBML: an API library for SBML". Bioinformatics 24 (6): 880-881. doi:10.1093/bioinformatics/btn051. PMC PMC2517632. PMID 18252737. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2517632.
- ↑ Le Hors, A.; Le Hégaret, P.; Wood, L. et al. (2004). "Document Object Model (DOM) Level 3 Core Specification". W3C. https://www.w3.org/TR/DOM-Level-3-Core/. Retrieved 14 March 2016.
- ↑ Hedengren, J.D.; Asgharzadeh, S.R.; Powell, K.M.; Edgar, T.F. (2014). "Nonlinear Modeling, Estimation and Predictive Control in APMonitor". Computers & Chemical Engineering 70: 133–148. doi:10.1016/j.compchemeng.2014.04.013.
- ↑ Demir, E.; Cary, M.P.; Paley, S. et al. (2010). "The BioPAX community standard for pathway data sharing". Nature Biotechnology 28 (9): 935–942. doi:10.1038/nbt.1666.
- ↑ Gansner, E.R.; North, S.C. (2000). "An open graph visualization system and its applications to software engineering". Software: Practice and Experience 30 (11): 1203–1233. doi:10.1002/1097-024X(200009)30:11<1203::AID-SPE338>3.0.CO;2-N.
- ↑ Van Iersel, M.P.; Kelder, T.; Pico, A.R. et al. (2008). "Presenting and exploring biological pathways with PathVisio". BMC Bioinformatics 9: 399. doi:10.1186/1471-2105-9-399. PMC PMC2569944. PMID 18817533. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2569944.
- ↑ Pico, A.R.; Kelder, T.; Van Iersel, M.P. et al. (2008). "WikiPathways: Pathway editing for the people". PLoS Biology 6 (7): e184. doi:10.1371/journal.pbio.0060184. PMC PMC2475545. PMID 18651794. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2475545.
- ↑ Stiles, J.R.; Bartol, T.M. (2000). "Chapter 4. Monte Carlod Methods for Simulating Realistic Synaptic Microphysiology Using MCell". In De Schutter, E.. Computational Neuroscience: Realistic Modeling for Experimentalists. CRC Press. doi:10.1201/9781420039290.ch4. ISBN 9781420039290.
- ↑ Eaton, J.W. (2015). "GNU Octave". Free Software Foundation, Inc. http://www.gnu.org/software/octave/doc/interpreter/. Retrieved 14 March 2016.
- ↑ Le Novère, N.; Hucka, M.; Mi, H. et al. (2009). "The Systems Biology Graphical Notation". Nature Biotechnology 27 (8): 735-741. doi:10.1038/nbt.1558. PMID 19668183.
- ↑ Ermentrout, B. (2002). Simulating, Analyzing, and Animating Dynamical Systems: A Guide to XPPAUT for Researchers and Students. Society for Industrial and Applied Mathematics. pp. 298. doi:10.1137/1.9780898718195. ISBN 9780898718195.
- ↑ Le Novère, N.; Bornstein, B.; Broicher, A. et al. (2006). "BioModels Database: A free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems". Nucleic Acids Research 34 (Suppl 1): D689-D691. doi:10.1093/nar/gkj092. PMC PMC1347454. PMID 16381960. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1347454.
- ↑ Le Novère, N.; Finney, A.; Hucka, M. et al. (2005). "Minimum Information Required In the Annotation of Models (MIRIAM)". Nature Biotechnology 23 (12): 1509-1515. doi:10.1038/nbt1156. PMID 16333295.
- ↑ "System Biology Format Converter (SBFC) Online". European Bioinformatics Institute. http://www.ebi.ac.uk/biomodels/tools/converters/. Retrieved 14 March 2016.
- ↑ "Roadmap". Systems Biology Format Converter (SBFC). SourceForge. http://sbfc.sourceforge.net/mediawiki/index.php/Roadmap. Retrieved 14 March 2016.
- ↑ "OSGi: The Dynamic Module System for Java". OSGi Alliance. https://www.osgi.org/. Retrieved 14 March 2016.
- ↑ Le Novère, N.; Hucka, M.; Mi, H. et al. (2003). "Cytoscape: A software environment for integrated models of biomolecular interaction networks". Nature Biotechnology 13 (11): 2498-2504. doi:10.1101/gr.1239303. PMC PMC403769. PMID 14597658. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC403769.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.