Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Grimes BMCBioinformatics2014 15.jpg|220px]]</div> | ||
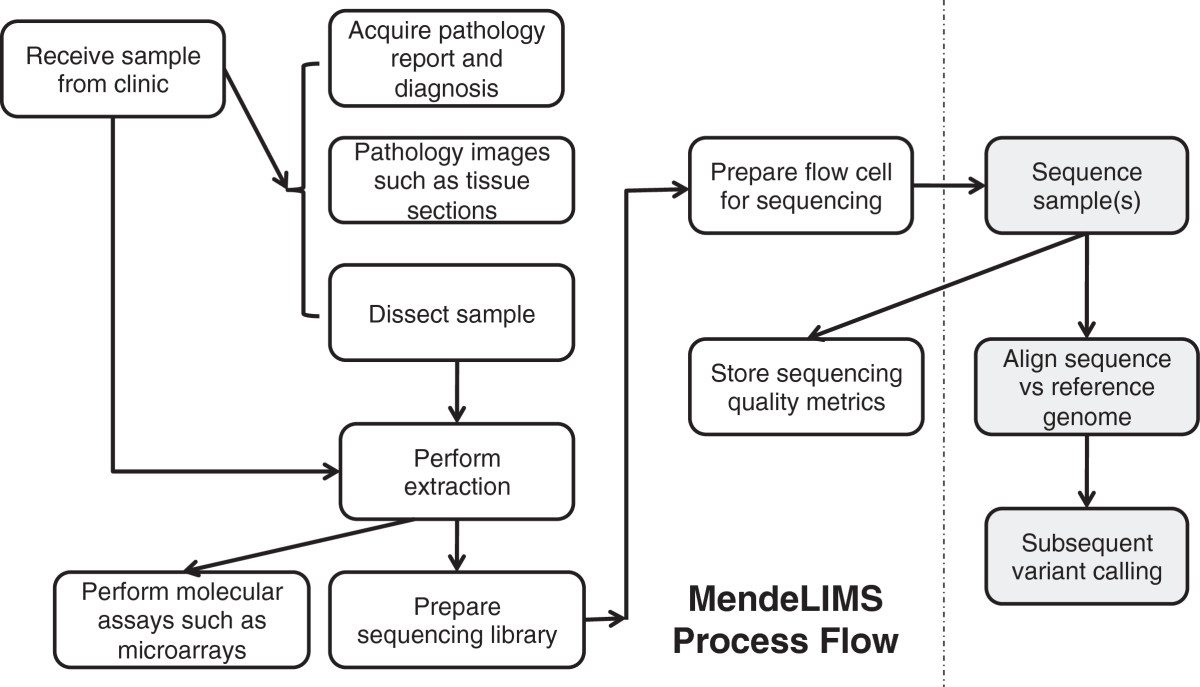
'''"[[Journal: | '''"[[Journal:MendeLIMS: A web-based laboratory information management system for clinical genome sequencing|MendeLIMS: A web-based laboratory information management system for clinical genome sequencing]]"''' | ||
Large clinical genomics studies using next generation DNA sequencing require the ability to select and track samples from a large population of patients through many experimental steps. With the number of clinical genome sequencing studies increasing, it is critical to maintain adequate [[laboratory information management system]]s to manage the thousands of patient samples that are subject to this type of genetic analysis. | |||
To meet the needs of clinical population studies using genome sequencing, we developed a web-based laboratory information management system (LIMS) with a flexible configuration that is adaptable to continuously evolving experimental protocols of next generation DNA sequencing technologies. Our system is referred to as [[Stanford University School of Medicine#MendeLIMS|MendeLIMS]], is easily implemented with open source tools and is also highly configurable and extensible. MendeLIMS has been invaluable in the management of our clinical genome sequencing studies. ('''[[Journal:MendeLIMS: A web-based laboratory information management system for clinical genome sequencing|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': [[Journal:Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations|Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations]], [[Journal:Requirements for data integration platforms in biomedical research networks: A reference model|Requirements for data integration platforms in biomedical research networks: A reference model | ''Recently featured'': [[Journal:Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages|Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages]], [[Journal:Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations|Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations]], [[Journal:Requirements for data integration platforms in biomedical research networks: A reference model|Requirements for data integration platforms in biomedical research networks: A reference model]] | ||
Revision as of 19:08, 5 January 2016
"MendeLIMS: A web-based laboratory information management system for clinical genome sequencing"
Large clinical genomics studies using next generation DNA sequencing require the ability to select and track samples from a large population of patients through many experimental steps. With the number of clinical genome sequencing studies increasing, it is critical to maintain adequate laboratory information management systems to manage the thousands of patient samples that are subject to this type of genetic analysis.
To meet the needs of clinical population studies using genome sequencing, we developed a web-based laboratory information management system (LIMS) with a flexible configuration that is adaptable to continuously evolving experimental protocols of next generation DNA sequencing technologies. Our system is referred to as MendeLIMS, is easily implemented with open source tools and is also highly configurable and extensible. MendeLIMS has been invaluable in the management of our clinical genome sequencing studies. (Full article...)
Recently featured: Personalized Oncology Suite: Integrating next-generation sequencing data and whole-slide bioimages, Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations, Requirements for data integration platforms in biomedical research networks: A reference model