File:Fig10 Garza BMCBioinformatics2016 17.gif
Fig10_Garza_BMCBioinformatics2016_17.gif (777 × 580 pixels, file size: 156 KB, MIME type: image/gif)
Summary
| Description |
Fig. 10 Label-free quantification pipeline as imported from the KNIME Analytics Platform into gUSE using the KNIME2gUSE extension. Note how parameter seep elements depicted in Fig. 9 such as ZipLoopStart and ZipLoopEnd are not present in gUSE. This is due to the fact that gUSE implements parameter seep by setting properties in input and output ports of the corresponding nodes. |
|---|---|
| Source |
de la Garza, L.; Veit, J.; Szolek, A.; Röttig, M.; Aiche, S.; Gesing, S.; Reinert, K.; Kohlbacher, O. (2016). "From the desktop to the grid: Scalable bioinformatics via workflow conversion". BMC Bioinformatics 17: 127. doi:10.1186/s12859-016-0978-9. PMC PMC4788856. PMID 26968893. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4788856. |
| Date |
2016 |
| Author |
de la Garza, L.; Veit, J.; Szolek, A.; Röttig, M.; Aiche, S.; Gesing, S.; Reinert, K.; Kohlbacher, O. |
| Permission (Reusing this file) |
|
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution 4.0 License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 17:43, 14 June 2016 | 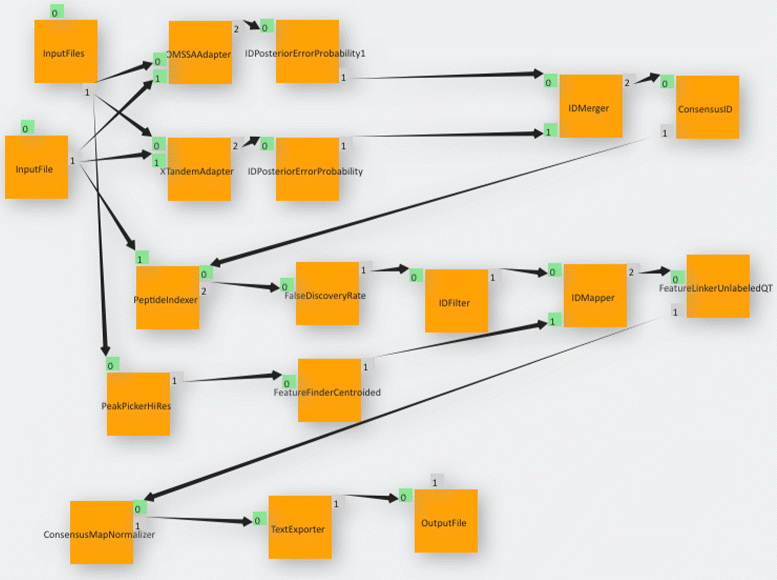 | 777 × 580 (156 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file:









