Difference between revisions of "File:Fig1 Barker BMCBio2013 14.jpg"
Shawndouglas (talk | contribs) |
Shawndouglas (talk | contribs) (Added summary.) |
||
| Line 1: | Line 1: | ||
{{Information | |||
|Description='''Figure 1. 4273''π'' screenshot.''' Practical, Week 7. Predicted gene structures of the Or98b gene in the Drosophila sechellia genome are being compared. Different predictions are made de novo by SNAP, run in a terminal on the Raspberry Pi; GeneWise, run online at the European Bioinformatics Institute, with the D. melanogaster protein being aligned against D. sechellia genomic DNA; and using TBLASTN, run online at FlyBase. FlyBase also displays the annotation of this gene from its database. This case study arose during research for a comparative study of the chemoreceptor superfamily in Drosophila. The 4273''π'' background desktop image includes hand-written versions of material from 4273''π'' Bioinformatics for Biologists and from the module BL3320 Practical Statistics for Biologists, which most students at the University of St Andrews would have taken earlier. It also includes a copy of one of the first published phylogenetic trees, from Vestiges of the Natural History of Creation, written in St Andrews. | |||
|Source={{cite journal |url=http://www.biomedcentral.com/1471-2105/14/243 |title=4273''π'': Bioinformatics education on low cost ARM hardware |journal=BMC Bioinformatics |author=Barker, Daniel; Ferrier, David E.K.; Holland, Peter W.H.; Mitchell, John B.O.; Plaisier, Heleen; Ritchie, Michael G.; Smart, Steven D. |volume=14 |pages=243 |year=2013 |doi=10.1186/1471-2105-14-243 |issn=1471-2105}} | |||
|Author=Barker, Daniel; Ferrier, David E.K.; Holland, Peter W.H.; Mitchell, John B.O.; Plaisier, Heleen; Ritchie, Michael G.; Smart, Steven D. | |||
|Date=2015 | |||
|Permission=[http://creativecommons.org/licenses/by/2.0/ Creative Commons Attribution 2.0 Generic] | |||
}} | |||
== Licensing == | == Licensing == | ||
{{cc-by-2.0}} | {{cc-by-2.0}} | ||
Latest revision as of 18:16, 14 November 2015
| Description |
Figure 1. 4273π screenshot. Practical, Week 7. Predicted gene structures of the Or98b gene in the Drosophila sechellia genome are being compared. Different predictions are made de novo by SNAP, run in a terminal on the Raspberry Pi; GeneWise, run online at the European Bioinformatics Institute, with the D. melanogaster protein being aligned against D. sechellia genomic DNA; and using TBLASTN, run online at FlyBase. FlyBase also displays the annotation of this gene from its database. This case study arose during research for a comparative study of the chemoreceptor superfamily in Drosophila. The 4273π background desktop image includes hand-written versions of material from 4273π Bioinformatics for Biologists and from the module BL3320 Practical Statistics for Biologists, which most students at the University of St Andrews would have taken earlier. It also includes a copy of one of the first published phylogenetic trees, from Vestiges of the Natural History of Creation, written in St Andrews. |
|---|---|
| Source |
Barker, Daniel; Ferrier, David E.K.; Holland, Peter W.H.; Mitchell, John B.O.; Plaisier, Heleen; Ritchie, Michael G.; Smart, Steven D. (2013). "4273π: Bioinformatics education on low cost ARM hardware". BMC Bioinformatics 14: 243. doi:10.1186/1471-2105-14-243. ISSN 1471-2105. http://www.biomedcentral.com/1471-2105/14/243. |
| Date |
2015 |
| Author |
Barker, Daniel; Ferrier, David E.K.; Holland, Peter W.H.; Mitchell, John B.O.; Plaisier, Heleen; Ritchie, Michael G.; Smart, Steven D. |
| Permission (Reusing this file) |
|
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution 2.0 License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 18:10, 14 November 2015 | 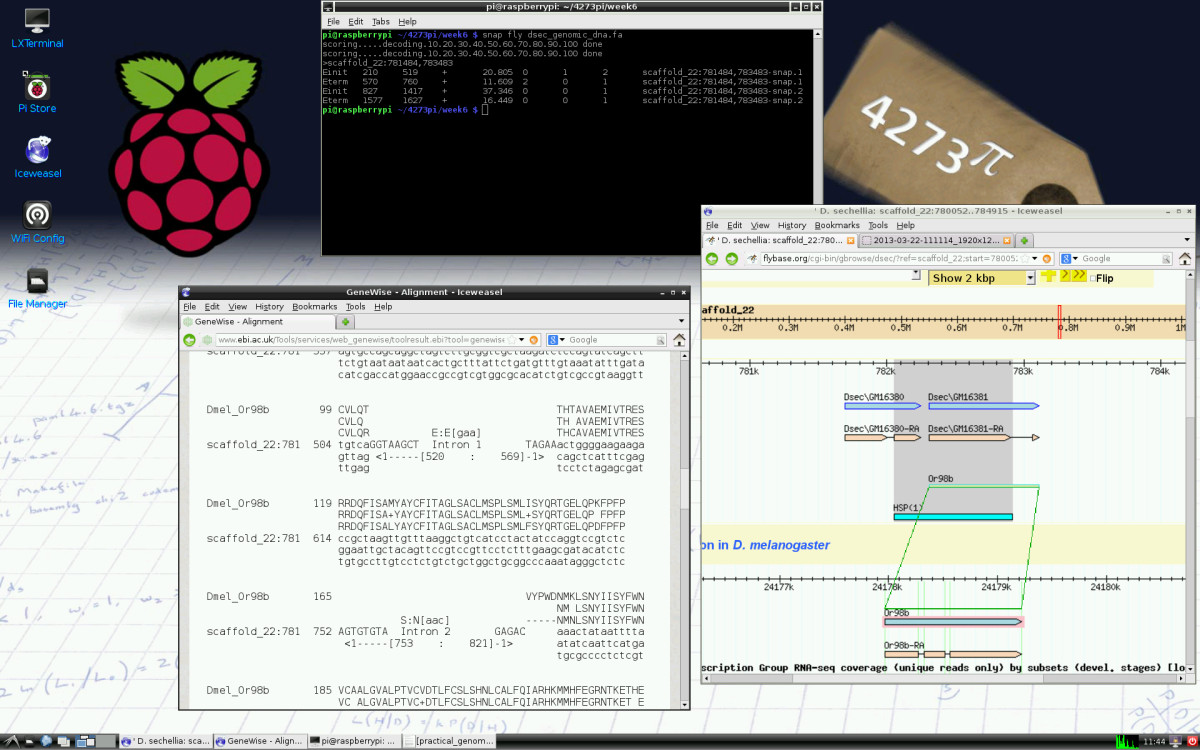 | 1,200 × 750 (216 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following 2 pages use this file:









