Difference between revisions of "File:Fig2 OConnor BMCInformatics2010 11-12.jpg"
Shawndouglas (talk | contribs) |
Shawndouglas (talk | contribs) (Added summary.) |
||
| Line 1: | Line 1: | ||
==Summary== | |||
{{Information | |||
|Description='''Figure 2. Load and query performance.''' Comparisons of load and query times between the HBase and BerkeleyDB backend. (a) Load times for the “1102 GBM” tumor/normal genomes where compared between HBase and BerkeleyDB. Both used a single-threaded approach to better compare relative performance. Both perform similarly but over time the load times for BerkeleyDB increase faster than with HBase. (b) Comparison of querying the 1102 genome database between BerkeleyDB, HBase single threaded, and HBase using MapReduce. Beyond 3M variants BerkeleyDB query times increase dramatically while both query types for HBase perform linearly, with MapReduce consistently exhibiting the best performance. | |||
|Source={{cite journal |url=http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-11-S12-S2 |title=SeqWare Query Engine: Storing and searching sequence data in the cloud |journal=BMC Informatics |author=O’Connor, Brian D.; Merriman, Barry; Nelson, Stanley F. |volume=11 |issue=12 |pages=S2 |year=2010 |doi=10.1186/1471-2105-11-S12-S2 |issn=1471-2105}} | |||
|Author=O’Connor, Brian D.; Merriman, Barry; Nelson, Stanley F. | |||
|Date=2010 | |||
|Permission=[http://creativecommons.org/licenses/by/2.0 Creative Commons Attribution 2.0 Generic] | |||
}} | |||
== Licensing == | == Licensing == | ||
{{cc-by-2.0}} | {{cc-by-2.0}} | ||
Latest revision as of 19:42, 28 December 2015
Summary
| Description |
Figure 2. Load and query performance. Comparisons of load and query times between the HBase and BerkeleyDB backend. (a) Load times for the “1102 GBM” tumor/normal genomes where compared between HBase and BerkeleyDB. Both used a single-threaded approach to better compare relative performance. Both perform similarly but over time the load times for BerkeleyDB increase faster than with HBase. (b) Comparison of querying the 1102 genome database between BerkeleyDB, HBase single threaded, and HBase using MapReduce. Beyond 3M variants BerkeleyDB query times increase dramatically while both query types for HBase perform linearly, with MapReduce consistently exhibiting the best performance. |
|---|---|
| Source |
O’Connor, Brian D.; Merriman, Barry; Nelson, Stanley F. (2010). "SeqWare Query Engine: Storing and searching sequence data in the cloud". BMC Informatics 11 (12): S2. doi:10.1186/1471-2105-11-S12-S2. ISSN 1471-2105. http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-11-S12-S2. |
| Date |
2010 |
| Author |
O’Connor, Brian D.; Merriman, Barry; Nelson, Stanley F. |
| Permission (Reusing this file) |
|
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution 2.0 License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 19:41, 28 December 2015 | 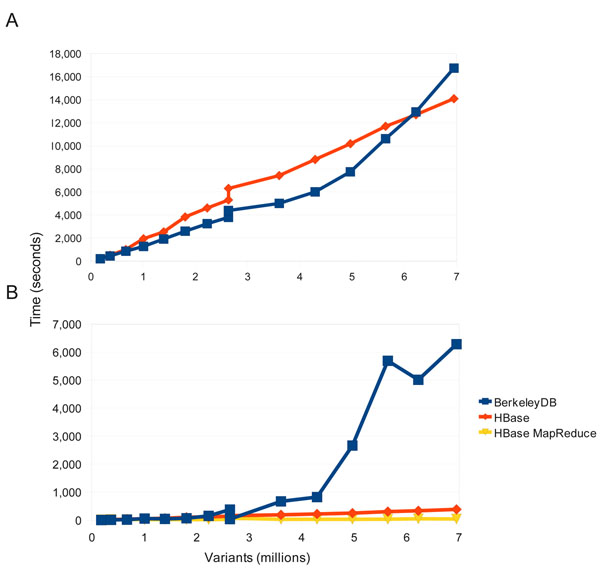 | 600 × 570 (59 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file:









