Difference between revisions of "File:Fig4 Ogle FrontBigData2021 4.jpg"
Shawndouglas (talk | contribs) |
Shawndouglas (talk | contribs) (Added summary.) |
||
| Line 1: | Line 1: | ||
==Summary== | |||
{{Information | |||
|Description='''Figure 4.''' The effect of NDN caching on DNA dataset insertion into GEMmaker workflow. A set of three NCBI SRA ''Arabidopsis thaliana'' datasets (SRR5263229:167.9 Mb, SRR5263230:167.8 Mb, SRR5263231:167.9 Mb) was sequentially transferred three times (black bars [transfer I], gray bars [transfer II], white bars [transfer III]) for insertion into the GEMmaker workflow from the NDN testbed or from the SRA repository over the internet via HTTP. The y-axis in both panels shows the transfer time of all three datasets, from request to workflow accessibility, in seconds. Error bars represent standard error of the mean. ('''A''') The aggregate transfer times are shown for the three sequential transfers with HTTP, NDN tested without caching, and NDN with caching at 655,360 packets. ('''B''') The effect of varying cache size (packet number in parentheses) is shown. The x-axis shows the cache capacity in packet numbers. | |||
|Source={{cite journal |title=Named data networking for genomics data management and integrated workflows |journal=Frontiers in Big Data |author=Ogle, C.; Reddick, D.; McKnight, C.; Biggs, T.; Pauly, R.; Ficklin, S.P.; Feltus, F.A.; Shannigrahi, S. |volume=4 |at=582468 |year=2021 |doi=10.3389/fdata.2021.582468}} | |||
|Author=Ogle, C.; Reddick, D.; McKnight, C.; Biggs, T.; Pauly, R.; Ficklin, S.P.; Feltus, F.A.; Shannigrahi, S. | |||
|Date=2021 | |||
|Permission=[http://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International] | |||
}} | |||
== Licensing == | == Licensing == | ||
{{cc-by-4.0}} | {{cc-by-4.0}} | ||
Latest revision as of 22:30, 19 March 2021
Summary
| Description |
Figure 4. The effect of NDN caching on DNA dataset insertion into GEMmaker workflow. A set of three NCBI SRA Arabidopsis thaliana datasets (SRR5263229:167.9 Mb, SRR5263230:167.8 Mb, SRR5263231:167.9 Mb) was sequentially transferred three times (black bars [transfer I], gray bars [transfer II], white bars [transfer III]) for insertion into the GEMmaker workflow from the NDN testbed or from the SRA repository over the internet via HTTP. The y-axis in both panels shows the transfer time of all three datasets, from request to workflow accessibility, in seconds. Error bars represent standard error of the mean. (A) The aggregate transfer times are shown for the three sequential transfers with HTTP, NDN tested without caching, and NDN with caching at 655,360 packets. (B) The effect of varying cache size (packet number in parentheses) is shown. The x-axis shows the cache capacity in packet numbers. |
|---|---|
| Source |
Ogle, C.; Reddick, D.; McKnight, C.; Biggs, T.; Pauly, R.; Ficklin, S.P.; Feltus, F.A.; Shannigrahi, S. (2021). "Named data networking for genomics data management and integrated workflows". Frontiers in Big Data 4: 582468. doi:10.3389/fdata.2021.582468. |
| Date |
2021 |
| Author |
Ogle, C.; Reddick, D.; McKnight, C.; Biggs, T.; Pauly, R.; Ficklin, S.P.; Feltus, F.A.; Shannigrahi, S. |
| Permission (Reusing this file) |
|
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution 4.0 License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 22:28, 19 March 2021 | 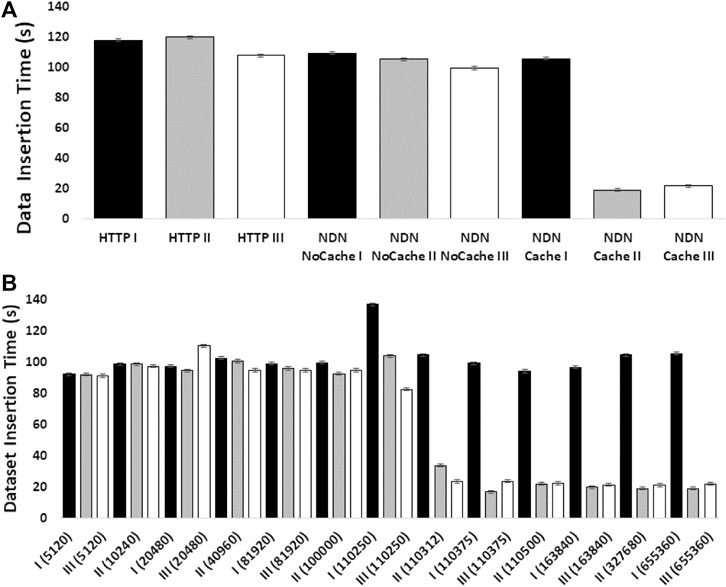 | 726 × 587 (58 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file:









