Difference between revisions of "Journal:A polyglot approach to bioinformatics data integration: A phylogenetic analysis of HIV-1"
Shawndouglas (talk | contribs) (Added content. Saving and adding more.) |
Shawndouglas (talk | contribs) (Added content. Saving and adding more.) |
||
| Line 33: | Line 33: | ||
Molecular biology is now plagued with the challenges facing numerous other fields – big data. Cloud-based solutions, e.g. CloudBurst<ref name="SchatzCloud09">{{cite journal |title=CloudBurst: Highly sensitive read mapping with MapReduce |journal=Bioinformatics |author=Schatz, M.C. |volume=25 |issue=11 |pages=1363-9 |year=2009 |doi=10.1093/bioinformatics/btp236 |pmid=19357099 |pmc=PMC2682523}}</ref>, Atlas2<ref name="EvaniAtlas12">{{cite journal |title=Atlas2 Cloud: A framework for personal genome analysis in the cloud |journal=BMC Genomics |author=Evani, U.S.; Challis, D.; Yu, J. et al. |volume=13 |issue=Suppl 6 |pages=S19 |year=2012 |doi=10.1186/1471-2164-13-S6-S19 |pmid=23134663 |pmc=PMC3481437}}</ref>, and Rainbow<ref name="ZhaoRainbow13">{{cite journal |title=Rainbow: A tool for large-scale whole-genome sequencing data analysis using cloud computing |journal=BMC Genomics |author=Zhao, S.; Prenger, K.; Smith, L. et al. |volume=14 |pages=425 |year=2013 |doi=10.1186/1471-2164-14-425 |pmid=23802613 |pmc=PMC3698007}}</ref>, have provided much needed leverage to meet these demands, facilitating large-scale sequence analyses, while also introducing new difficulties.<ref name="SchatzCloud10">{{cite journal |title=Cloud computing and the DNA data race |journal=Nature Biotechnology |author=Schatz, M.C.; Langmead, B.; Salzberg, S.L. |volume=28 |pages=7 |year=2010 |doi=10.1038/nbt0710-691 |pmid=20622843 |pmc=PMC2904649}}</ref> Furthermore, noSQL databases afford a streamlined solution to both manage large datasets and simplify data retrieval and subsequent analysis. The added benefit of agility and scalability of noSQL databases is ideal for the rapidly advancing trends in DNA sequencing technologies, and it is thus not surprising that noSQL databases have been gaining traction in molecular studies.<ref name="BorozanCaPSID12">{{cite journal |title=CaPSID: A bioinformatics platform for computational pathogen sequence identification in human genomes and transcriptomes |journal=BMC Bioinformatics |author=Borozan, I.; Wilson, S.; Blanchette, P. |volume=13 |pages=206 |year=2012 |doi=10.1186/1471-2105-13-206 |pmid=22901030 |pmc=PMC3464663}}</ref><ref name="Hird_lociNGS12">{{cite journal |title=lociNGS: A lightweight alternative for assessing suitability of next-generation loci for evolutionary analysis |journal=PLoS One |author=Hird, S.M. |volume=7 |issue=10 |pages=e46847 |year=2012 |doi=10.1371/journal.pone.0046847 |pmid=23071651 |pmc=PMC3468592}}</ref><ref name="NingthoujamNoSQL14">{{cite journal |title=NoSQL data model for semi-automatic integration of ethnomedicinal plant data from multiple sources |journal=Phytochemical Analysis |author=Ningthoujam, S.S.; Choudhury, M.D.; Potsangbam, K.S. et al. |volume=25 |issue=6 |pages=495-507 |year=2014 |doi=10.1002/pca.2520 |pmid=24737485}}</ref> | Molecular biology is now plagued with the challenges facing numerous other fields – big data. Cloud-based solutions, e.g. CloudBurst<ref name="SchatzCloud09">{{cite journal |title=CloudBurst: Highly sensitive read mapping with MapReduce |journal=Bioinformatics |author=Schatz, M.C. |volume=25 |issue=11 |pages=1363-9 |year=2009 |doi=10.1093/bioinformatics/btp236 |pmid=19357099 |pmc=PMC2682523}}</ref>, Atlas2<ref name="EvaniAtlas12">{{cite journal |title=Atlas2 Cloud: A framework for personal genome analysis in the cloud |journal=BMC Genomics |author=Evani, U.S.; Challis, D.; Yu, J. et al. |volume=13 |issue=Suppl 6 |pages=S19 |year=2012 |doi=10.1186/1471-2164-13-S6-S19 |pmid=23134663 |pmc=PMC3481437}}</ref>, and Rainbow<ref name="ZhaoRainbow13">{{cite journal |title=Rainbow: A tool for large-scale whole-genome sequencing data analysis using cloud computing |journal=BMC Genomics |author=Zhao, S.; Prenger, K.; Smith, L. et al. |volume=14 |pages=425 |year=2013 |doi=10.1186/1471-2164-14-425 |pmid=23802613 |pmc=PMC3698007}}</ref>, have provided much needed leverage to meet these demands, facilitating large-scale sequence analyses, while also introducing new difficulties.<ref name="SchatzCloud10">{{cite journal |title=Cloud computing and the DNA data race |journal=Nature Biotechnology |author=Schatz, M.C.; Langmead, B.; Salzberg, S.L. |volume=28 |pages=7 |year=2010 |doi=10.1038/nbt0710-691 |pmid=20622843 |pmc=PMC2904649}}</ref> Furthermore, noSQL databases afford a streamlined solution to both manage large datasets and simplify data retrieval and subsequent analysis. The added benefit of agility and scalability of noSQL databases is ideal for the rapidly advancing trends in DNA sequencing technologies, and it is thus not surprising that noSQL databases have been gaining traction in molecular studies.<ref name="BorozanCaPSID12">{{cite journal |title=CaPSID: A bioinformatics platform for computational pathogen sequence identification in human genomes and transcriptomes |journal=BMC Bioinformatics |author=Borozan, I.; Wilson, S.; Blanchette, P. |volume=13 |pages=206 |year=2012 |doi=10.1186/1471-2105-13-206 |pmid=22901030 |pmc=PMC3464663}}</ref><ref name="Hird_lociNGS12">{{cite journal |title=lociNGS: A lightweight alternative for assessing suitability of next-generation loci for evolutionary analysis |journal=PLoS One |author=Hird, S.M. |volume=7 |issue=10 |pages=e46847 |year=2012 |doi=10.1371/journal.pone.0046847 |pmid=23071651 |pmc=PMC3468592}}</ref><ref name="NingthoujamNoSQL14">{{cite journal |title=NoSQL data model for semi-automatic integration of ethnomedicinal plant data from multiple sources |journal=Phytochemical Analysis |author=Ningthoujam, S.S.; Choudhury, M.D.; Potsangbam, K.S. et al. |volume=25 |issue=6 |pages=495-507 |year=2014 |doi=10.1002/pca.2520 |pmid=24737485}}</ref> | ||
With the increase in the amount of publicly available genomic sequence data, progress can be stymied by the simple task of collecting sequence data and associated, relevant metadata. In an effort to facilitate the aggregation and management of genomic data for subsequent analyses, we have developed a polyglot approach involving multiple languages (Python and Scala), libraries ([http://flask.pocoo.org Flask] and [http://biojava.org BioJavaX]), and persistence mechanisms (text files and [http://www.mongodb.org MongoDB] NoSQL databases). Individual genes or all genes for a given species can be examined beyond just the sequence itself, including information regarding, for instance, the location and date of isolation. | |||
The code developed is agile; it can be applied for any organism of interest to the user. The approach can be customized for any species of interest. The presented solution is developed with downstream evolutionary analyses in mind, as shown by a proof-of-concept study of the evolution of HIV. Our investigation into the three main HIV genes: gag, pol, and env, reveals spatial and temporal factors influence the evolution of the individual genes uniquely. The web service for the HIV collection (as well as other datasets investigated by the authors) is publicly available at http://hivdb.cs.luc.edu, and the scripts for generating such a data collection are publicly available at https://github.com/LoyolaChicagoCode/hiv-biojava-scala. | |||
==Results and discussion== | |||
===Data pipeline for collecting sequence data=== | |||
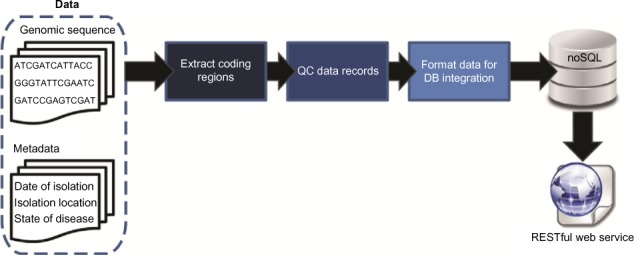
Code has been developed to aggregate genomic sequence data and available sequence metadata for subsequent analyses. Figure 1 summarizes this process. All complete and partial genome sequences were parsed and separated into individual folders for each gene via a Scala parser utilizing the [http://biojava.org BioJavaX] library. Each sequence was stored in its gene folder with any relevant metadata available within the GenBank file. The generated folders for each of the parsed genes were then pipelined through several python scripts in order to accomplish several post-processing tasks. First, duplicate gene sequences parsed from the same genome were removed. Second, the gene folders were used to create FASTA-formatted records for each of the gene sequences with any necessary metadata stored in the resulting record’s FASTA header. Finally, the [https://pypi.python.org/pypi/pymongo/ PyMongo library] was used to insert each of the final FASTA records within our publicly exposed MongoDB database. | |||
[[File:Fig1 Reisman EBioinformatics2016 12.jpg|750px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="750px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 1.''' Schematic of data pipeline and access</blockquote> | |||
|- | |||
|} | |||
|} | |||
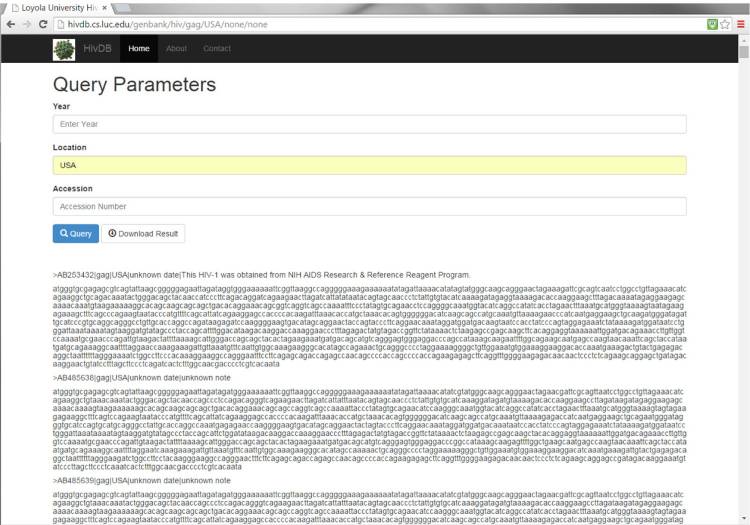
Genomic sequence data can then be accessed via a RESTful<ref name="FieldingArch00">{{cite web |url=https://www.ics.uci.edu/~fielding/pubs/dissertation/top.htm |title=Architectural Styles and the Design of Network-based Software Architectures |author=Fielding, R.T. |publisher=University of California, Irvine |date=2000}}</ref> web service located at http://hivdb.cs.luc.edu. This architecture allows our service to be easily and efficiently accessed by any future data consumers via common web protocols. Data can be queried based upon attributes regarding the source of the data. For example, as shown in Figure 2, the gag gene sequences from strains isolated in the USA can be accessed via the web service. The user can specify search criteria including a year (or range of years) of isolation, the location of isolation, and/or accession number. Sequences meeting the user’s search criteria can be returned to the web via the Query button or downloaded. All sequence results are in FASTA format for subsequent analysis, such as sequence alignment, primer development, and phylogenetic analysis. | |||
[[File:Fig2 Reisman EBioinformatics2016 12.jpg|630px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="630px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 2.''' Gene sequence data presented through RESTful web service. Users can query for specific information, eg, as shown here are HIV isolates from the USA, via the Query button or download sequence files in FASTA format meeting their search criteria.</blockquote> | |||
|- | |||
|} | |||
|} | |||
The pipeline has been developed to facilitate users to create repositories for an organism(s) of interest as well as queryable aspects of the sequence annotations. Users need only supply sequences and select attributes and/or genes of interest (otherwise all attributes and genes will be selected). Data are automatically processed. Furthermore, the pipeline is not restricted to publicly available data; any GenBank-formatted file (public or private) can be included. Given the increased throughput of contemporary sequencing technologies and the decreased cost in sequencing runs, whole genome sequencing is being conducted at unprecedented rates. As such, researchers sequencing novel strains or isolates can incorporate their strains into the data repository once GenBank files are generated. Although this pipeline has been employed by the authors for the analysis of several different taxa, the RESTful web service presented here includes publicly available data for HIV-1 sequences. | |||
===Case study: Investigation of the evolution of HIV-1=== | |||
All publicly available complete and near-complete HIV-1 genomic sequences, totaling more than 6,000 sequences, were retrieved from the National Center for Biotechnology Information (NCBI) and processed by our pipeline (see the "Methods and materials" section). Individual gene sequences are publicly available at http://hivdb.cs.luc.edu. Data are accessible in FASTA format to facilitate downstream analyses. To incorporate the metadata collected, including country and date of isolation, this information has been integrated into the FASTA record header. HIV-1 sequences were selected as a proof of concept for this tool as HIV sequences are among the most well curated, thanks in large part to efforts such as those at the Los Alamos National Laboratory’s [http://www.hiv.lanl.gov/ HIV sequence database]. | |||
Previously, phylogenetics has shed light on the origin of HIV and played a key role in identifying recombination events.<ref name="GaoOrigin99">{{cite journal |title=Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes |journal=Nature |author=Gao, F.; Bailes, E.; Robertson, D.L. et al. |volume=397 |issue=6718 |pages=436-41 |year=1999 |doi=10.1038/17130 |pmid=9989410}}</ref><ref name="GaoGenetic94">{{cite journal |title=Genetic diversity of human immunodeficiency virus type 2: Evidence for distinct sequence subtypes with differences in virus biology |journal=Journal of Virology |author=Gao, F.; Yue, L.; Robertson, D.L. et al. |volume=68 |issue=11 |pages=7433-47 |year=1994 |pmid=7933127 |pmc=PMC237186}}</ref> As previous molecular studies have shown, the evolutionary history of the HIV-1 lineage includes three groups (M, N, and O) representative of separate transfers from chimpanzees.<ref name="RambautTheCau04">{{cite journal |title=The causes and consequences of HIV evolution |journal=Nature Reviews Genetics |author=Rambaut, A.; Posada, D.; Crandall, K.A.; Holmes, E.C. |volume=5 |issue=1 |pages=52–61 |year=2004 |doi=10.1038/nrg1246 |pmid=14708016}}</ref> Focusing on the three HIV genes gag, pol, and env, the hivdb data repository was queried for coding regions isolated from the same country as well as globally over a particular time period. Host, immunological and antiretroviral drug selection pressures have shaped much of the diversity observed within these three genes.<ref name="BrennerPhylo13">{{cite journal |title=Phylogenetic inferences on HIV-1 transmission: Implications for the design of prevention and treatment interventions |journal=AIDS |author=Brenner, B.; Wainberg, M.A.; Roger, M. |volume=27 |issue=7 |pages=1045-57 |year=2013 |doi=10.1097/QAD.0b013e32835cffd9 |pmid=23902920 |pmc=PMC3786580}}</ref> For instance, the investigation of the HIV gag gene sequences from the USA (2,048 sequences: 1990–2011) and Thailand (872 sequences: 2000–2011) is shown in Figure 3A and B, respectively. The phylogenic trees derived for different geographic regions revealed different tree topologies as expected. Sequences isolated during 2005 in the USA exhibit significant sequence variation, including a number of sequences which are distinctly different from sequences isolated during any other year (Fig. 3A). These two gag trees reveal a general trend observed for other countries and other genes: sequences isolated during the same year do not necessarily group together or exhibit a ladder-like topology frequently observed for intra-host HIV phylogenies<ref name="ShankarappaConsist99">{{cite journal |title=Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection |journal=Journal of Virology |author=Shankarappa, R.; Margolick, J.B.; Gange, S.J. et al. |volume=73 |issue=12 |pages=10489-502 |year=1999 |pmid=10559367 |pmc=PMC113104}}</ref>; this is in concordance with previous HIV survey findings that multiple lineages coexist at any given time.<ref name="Castro-NallarTheEvo12">{{cite journal |title=The evolution of HIV: Inferences using phylogenetics |journal=Molecular Phylogenetics and Evolution |author=Castro-Nallar, E.; Pérez-Losada, M.; Burton, G.F.; Crandall, K.A. |volume=62 |issue=2 |pages=777-92 |year=2012 |doi=10.1016/j.ympev.2011.11.019 |pmid=22138161 |pmc=PMC3258026}}</ref> | |||
==References== | ==References== | ||
Revision as of 19:05, 5 May 2016
| Full article title | A polyglot approach to bioinformatics data integration: A phylogenetic analysis of HIV-1 |
|---|---|
| Journal | Evolutionary Bioinformatics |
| Author(s) | Reisman, S.; Hatzopoulos, T.; Läufer, K.; Thiruvathukal, G.K.; Putonti, C. |
| Author affiliation(s) | Loyola University Chicago |
| Primary contact | Email: cputonti@luc.edu |
| Year published | 2016 |
| Volume and issue | 12 |
| Page(s) | 23–27 |
| DOI | 10.4137/EBO.S32757 |
| ISSN | 1176-9343 |
| Distribution license | Creative Commons Attribution-NonCommercial 3.0 Unported |
| Website | http://www.la-press.com/ |
| Download | http://www.la-press.com/ (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
As sequencing technologies continue to drop in price and increase in throughput, new challenges emerge for the management and accessibility of genomic sequence data. We have developed a pipeline for facilitating the storage, retrieval, and subsequent analysis of molecular data, integrating both sequence and metadata. Taking a polyglot approach involving multiple languages, libraries, and persistence mechanisms, sequence data can be aggregated from publicly available and local repositories. Data are exposed in the form of a RESTful web service, formatted for easy querying, and retrieved for downstream analyses. As a proof of concept, we have developed a resource for annotated HIV-1 sequences. Phylogenetic analyses were conducted for >6,000 HIV-1 sequences revealing spatial and temporal factors influence the evolution of the individual genes uniquely. Nevertheless, signatures of origin can be extrapolated even despite increased globalization. The approach developed here can easily be customized for any species of interest.
Keywords: polyglot programming, RESTful web service, phylogenetics
Introduction
The increased throughput, coupled with reduced cost and time, of contemporary sequencing technologies has led to a surge in the number of publicly available, complete, annotated genomic sequences. For smaller viral species, it is now feasible to not only produce a single genome for a species but also capture the diversity present in an ecological niche, the focus of numerous metagenomic studies[1][2][3] as well as more targeted investigations.[4] Furthermore, next-generation sequencing technologies have tremendous potential for the future of diagnostics and subsequent treatment choices, particularly for viral infections.[5] The sensitivity of deep sequencing can capture even rare variants in mixed infections as well as quasispecies.[6][7][8][9][10][11] Investigation of the viable variations within a viral species not only provides insight into the evolutionary history of a species but also unveils putative avenues for targeted therapies, such as small interfering RNAs[12][13][14][15] and control strategies.
Molecular biology is now plagued with the challenges facing numerous other fields – big data. Cloud-based solutions, e.g. CloudBurst[16], Atlas2[17], and Rainbow[18], have provided much needed leverage to meet these demands, facilitating large-scale sequence analyses, while also introducing new difficulties.[19] Furthermore, noSQL databases afford a streamlined solution to both manage large datasets and simplify data retrieval and subsequent analysis. The added benefit of agility and scalability of noSQL databases is ideal for the rapidly advancing trends in DNA sequencing technologies, and it is thus not surprising that noSQL databases have been gaining traction in molecular studies.[20][21][22]
With the increase in the amount of publicly available genomic sequence data, progress can be stymied by the simple task of collecting sequence data and associated, relevant metadata. In an effort to facilitate the aggregation and management of genomic data for subsequent analyses, we have developed a polyglot approach involving multiple languages (Python and Scala), libraries (Flask and BioJavaX), and persistence mechanisms (text files and MongoDB NoSQL databases). Individual genes or all genes for a given species can be examined beyond just the sequence itself, including information regarding, for instance, the location and date of isolation.
The code developed is agile; it can be applied for any organism of interest to the user. The approach can be customized for any species of interest. The presented solution is developed with downstream evolutionary analyses in mind, as shown by a proof-of-concept study of the evolution of HIV. Our investigation into the three main HIV genes: gag, pol, and env, reveals spatial and temporal factors influence the evolution of the individual genes uniquely. The web service for the HIV collection (as well as other datasets investigated by the authors) is publicly available at http://hivdb.cs.luc.edu, and the scripts for generating such a data collection are publicly available at https://github.com/LoyolaChicagoCode/hiv-biojava-scala.
Results and discussion
Data pipeline for collecting sequence data
Code has been developed to aggregate genomic sequence data and available sequence metadata for subsequent analyses. Figure 1 summarizes this process. All complete and partial genome sequences were parsed and separated into individual folders for each gene via a Scala parser utilizing the BioJavaX library. Each sequence was stored in its gene folder with any relevant metadata available within the GenBank file. The generated folders for each of the parsed genes were then pipelined through several python scripts in order to accomplish several post-processing tasks. First, duplicate gene sequences parsed from the same genome were removed. Second, the gene folders were used to create FASTA-formatted records for each of the gene sequences with any necessary metadata stored in the resulting record’s FASTA header. Finally, the PyMongo library was used to insert each of the final FASTA records within our publicly exposed MongoDB database.
|
Genomic sequence data can then be accessed via a RESTful[23] web service located at http://hivdb.cs.luc.edu. This architecture allows our service to be easily and efficiently accessed by any future data consumers via common web protocols. Data can be queried based upon attributes regarding the source of the data. For example, as shown in Figure 2, the gag gene sequences from strains isolated in the USA can be accessed via the web service. The user can specify search criteria including a year (or range of years) of isolation, the location of isolation, and/or accession number. Sequences meeting the user’s search criteria can be returned to the web via the Query button or downloaded. All sequence results are in FASTA format for subsequent analysis, such as sequence alignment, primer development, and phylogenetic analysis.
|
The pipeline has been developed to facilitate users to create repositories for an organism(s) of interest as well as queryable aspects of the sequence annotations. Users need only supply sequences and select attributes and/or genes of interest (otherwise all attributes and genes will be selected). Data are automatically processed. Furthermore, the pipeline is not restricted to publicly available data; any GenBank-formatted file (public or private) can be included. Given the increased throughput of contemporary sequencing technologies and the decreased cost in sequencing runs, whole genome sequencing is being conducted at unprecedented rates. As such, researchers sequencing novel strains or isolates can incorporate their strains into the data repository once GenBank files are generated. Although this pipeline has been employed by the authors for the analysis of several different taxa, the RESTful web service presented here includes publicly available data for HIV-1 sequences.
Case study: Investigation of the evolution of HIV-1
All publicly available complete and near-complete HIV-1 genomic sequences, totaling more than 6,000 sequences, were retrieved from the National Center for Biotechnology Information (NCBI) and processed by our pipeline (see the "Methods and materials" section). Individual gene sequences are publicly available at http://hivdb.cs.luc.edu. Data are accessible in FASTA format to facilitate downstream analyses. To incorporate the metadata collected, including country and date of isolation, this information has been integrated into the FASTA record header. HIV-1 sequences were selected as a proof of concept for this tool as HIV sequences are among the most well curated, thanks in large part to efforts such as those at the Los Alamos National Laboratory’s HIV sequence database.
Previously, phylogenetics has shed light on the origin of HIV and played a key role in identifying recombination events.[24][25] As previous molecular studies have shown, the evolutionary history of the HIV-1 lineage includes three groups (M, N, and O) representative of separate transfers from chimpanzees.[26] Focusing on the three HIV genes gag, pol, and env, the hivdb data repository was queried for coding regions isolated from the same country as well as globally over a particular time period. Host, immunological and antiretroviral drug selection pressures have shaped much of the diversity observed within these three genes.[27] For instance, the investigation of the HIV gag gene sequences from the USA (2,048 sequences: 1990–2011) and Thailand (872 sequences: 2000–2011) is shown in Figure 3A and B, respectively. The phylogenic trees derived for different geographic regions revealed different tree topologies as expected. Sequences isolated during 2005 in the USA exhibit significant sequence variation, including a number of sequences which are distinctly different from sequences isolated during any other year (Fig. 3A). These two gag trees reveal a general trend observed for other countries and other genes: sequences isolated during the same year do not necessarily group together or exhibit a ladder-like topology frequently observed for intra-host HIV phylogenies[28]; this is in concordance with previous HIV survey findings that multiple lineages coexist at any given time.[29]
References
- ↑ Fierer, N.; Breitbart, M.; Nulton, J. et al. (2007). "Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil". Applied and Environmental Microbiology 73 (21): 7059-66. doi:10.1128/AEM.00358-07. PMC PMC2074941. PMID 17827313. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2074941.
- ↑ Holmfeldt, K.; Solonenko, N.; Shah, M. et al. (2013). "Twelve previously unknown phage genera are ubiquitous in global oceans". Proceedings of the National Academy of Sciences of the United States of America 110 (31): 12798-803. doi:10.1073/pnas.1305956110. PMC PMC3732932. PMID 23858439. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3732932.
- ↑ Hurwitz, B.L.; Sullivan, M.B. (2013). "The Pacific Ocean Virome (POV): A marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology". PLoS One 8 (2): e57355. doi:10.1371/journal.pone.0057355. PMC PMC3585363. PMID 23468974. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3585363.
- ↑ Deng, L.; Ignacio-Espinoza, J.C.; Gregory, A.C. (2014). "Viral tagging reveals discrete populations in Synechococcus viral genome sequence space". Nature 513 (7517): 242-5. doi:10.1038/nature13459. PMID 25043051.
- ↑ Barzon, L.; Lavezzo, E.; Militello, V. et al. (2011). "Applications of next-generation sequencing technologies to diagnostic virology". International Journal of Medical Sciences 12 (11): 7861-84. doi:10.3390/ijms12117861. PMC PMC3233444. PMID 22174638. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3233444.
- ↑ Wang, C.; Mitsuya, Y.; Gharizadeh, B. et al. (2007). "Characterization of mutation spectra with ultra-deep pyrosequencing: Application to HIV-1 drug resistance". Genome Research 17 (8): 1195-201. doi:10.1101/gr.6468307. PMC PMC1933516. PMID 17600086. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1933516.
- ↑ Ramakrishnan, M.A.; Tu, Z.J.; Singh, S. et al. (2009). "The feasibility of using high resolution genome sequencing of influenza A viruses to detect mixed infections and quasispecies". PLoS One 4 (9): e7105. doi:10.1371/journal.pone.0007105. PMC PMC2740821. PMID 19771155. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2740821.
- ↑ Solmone, M.; Vincenti, D.; Prosperi, M.C. et al. (2009). "Use of massively parallel ultradeep pyrosequencing to characterize the genetic diversity of hepatitis B virus in drug-resistant and drug-naive patients and to detect minor variants in reverse transcriptase and hepatitis B S antigen". Journal of Virology 83 (4): 1718-26. doi:10.1128/JVI.02011-08. PMC PMC2643754. PMID 19073746. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2643754.
- ↑ Abdelrahman, T.; Hughes, J.; Main, J. et al. (2015). "Next-generation sequencing sheds light on the natural history of hepatitis C infection in patients who fail treatment". Hepatology 61 (1): 88–97. doi:10.1002/hep.27192. PMC PMC4303934. PMID 24797101. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4303934.
- ↑ Verheyen, J.; Litau, E.; Sing, T. et al. (2006). "Compensatory mutations at the HIV cleavage sites p7/p1 and p1/p6-gag in therapy-naive and therapy-experienced patients". Antiviral Therapy 11 (7): 879-87. PMID 17302250.
- ↑ Quiñones-Mateu, M.E.; Avila, S.; Reyes-Teran, G.; Martinez, M.A. (2015). "Deep sequencing: becoming a critical tool in clinical virology". Journal of Clinical Virology 61 (1): 9–19. doi:10.1016/j.jcv.2014.06.013. PMC PMC4119849. PMID 24998424. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4119849.
- ↑ Lares, M.R.; Rossi, J.J.; Ouellet, D.L. (2010). "RNAi and small interfering RNAs in human disease therapeutic applications". Trends in Biotechnology 28 (11): 570-9. doi:10.1016/j.tibtech.2010.07.009. PMC PMC2955826. PMID 20833440. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2955826.
- ↑ Truong, N.P.; Gu, W.; Prasadam, I. et al. (2013). "An influenza virus-inspired polymer system for the timed release of siRNA". Nature Communications 4: 1902. doi:10.1038/ncomms2905. PMID 23695696.
- ↑ Paul, A.M.; Shi, Y.; Acharya, D. et al. (2014). "Delivery of antiviral small interfering RNA with gold nanoparticles inhibits dengue virus infection in vitro". Journal of General Virology 95 (Pt 8): 1712-22. doi:10.1099/vir.0.066084-0. PMC PMC4103068. PMID 24828333. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4103068.
- ↑ Jin, F.; Li, S.; Zheng, K. et al. (2014). "Silencing herpes simplex virus type 1 capsid protein encoding genes by siRNA: A promising antiviral therapeutic approach". PLoS One 9 (5): e96623. doi:10.1371/journal.pone.0096623. PMC PMC4008601. PMID 24794394. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4008601.
- ↑ Schatz, M.C. (2009). "CloudBurst: Highly sensitive read mapping with MapReduce". Bioinformatics 25 (11): 1363-9. doi:10.1093/bioinformatics/btp236. PMC PMC2682523. PMID 19357099. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2682523.
- ↑ Evani, U.S.; Challis, D.; Yu, J. et al. (2012). "Atlas2 Cloud: A framework for personal genome analysis in the cloud". BMC Genomics 13 (Suppl 6): S19. doi:10.1186/1471-2164-13-S6-S19. PMC PMC3481437. PMID 23134663. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3481437.
- ↑ Zhao, S.; Prenger, K.; Smith, L. et al. (2013). "Rainbow: A tool for large-scale whole-genome sequencing data analysis using cloud computing". BMC Genomics 14: 425. doi:10.1186/1471-2164-14-425. PMC PMC3698007. PMID 23802613. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3698007.
- ↑ Schatz, M.C.; Langmead, B.; Salzberg, S.L. (2010). "Cloud computing and the DNA data race". Nature Biotechnology 28: 7. doi:10.1038/nbt0710-691. PMC PMC2904649. PMID 20622843. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2904649.
- ↑ Borozan, I.; Wilson, S.; Blanchette, P. (2012). "CaPSID: A bioinformatics platform for computational pathogen sequence identification in human genomes and transcriptomes". BMC Bioinformatics 13: 206. doi:10.1186/1471-2105-13-206. PMC PMC3464663. PMID 22901030. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3464663.
- ↑ Hird, S.M. (2012). "lociNGS: A lightweight alternative for assessing suitability of next-generation loci for evolutionary analysis". PLoS One 7 (10): e46847. doi:10.1371/journal.pone.0046847. PMC PMC3468592. PMID 23071651. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3468592.
- ↑ Ningthoujam, S.S.; Choudhury, M.D.; Potsangbam, K.S. et al. (2014). "NoSQL data model for semi-automatic integration of ethnomedicinal plant data from multiple sources". Phytochemical Analysis 25 (6): 495-507. doi:10.1002/pca.2520. PMID 24737485.
- ↑ Fielding, R.T. (2000). "Architectural Styles and the Design of Network-based Software Architectures". University of California, Irvine. https://www.ics.uci.edu/~fielding/pubs/dissertation/top.htm.
- ↑ Gao, F.; Bailes, E.; Robertson, D.L. et al. (1999). "Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes". Nature 397 (6718): 436-41. doi:10.1038/17130. PMID 9989410.
- ↑ Gao, F.; Yue, L.; Robertson, D.L. et al. (1994). "Genetic diversity of human immunodeficiency virus type 2: Evidence for distinct sequence subtypes with differences in virus biology". Journal of Virology 68 (11): 7433-47. PMC PMC237186. PMID 7933127. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC237186.
- ↑ Rambaut, A.; Posada, D.; Crandall, K.A.; Holmes, E.C. (2004). "The causes and consequences of HIV evolution". Nature Reviews Genetics 5 (1): 52–61. doi:10.1038/nrg1246. PMID 14708016.
- ↑ Brenner, B.; Wainberg, M.A.; Roger, M. (2013). "Phylogenetic inferences on HIV-1 transmission: Implications for the design of prevention and treatment interventions". AIDS 27 (7): 1045-57. doi:10.1097/QAD.0b013e32835cffd9. PMC PMC3786580. PMID 23902920. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3786580.
- ↑ Shankarappa, R.; Margolick, J.B.; Gange, S.J. et al. (1999). "Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection". Journal of Virology 73 (12): 10489-502. PMC PMC113104. PMID 10559367. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC113104.
- ↑ Castro-Nallar, E.; Pérez-Losada, M.; Burton, G.F.; Crandall, K.A. (2012). "The evolution of HIV: Inferences using phylogenetics". Molecular Phylogenetics and Evolution 62 (2): 777-92. doi:10.1016/j.ympev.2011.11.019. PMC PMC3258026. PMID 22138161. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3258026.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.