Journal:Developing a framework for open and FAIR data management practices for next generation risk- and benefit assessment of fish and seafood
| Full article title | Developing a framework for open and FAIR data management practices for next generation risk- and benefit assessment of fish and seafood |
|---|---|
| Journal | EFSA Journal |
| Author(s) | Pineda-Pampliega, Javier; Bernhard, Annette; Hannisdal, Rita; Ørnsrud, Robin; Mathisen, Gro H.; Solstad, Gisle; Rasinger, Josef D. |
| Author affiliation(s) | Norwegian Scientific Committee for Food and Environment |
| Primary contact | eu dash fora at efsa dot europa dot eu |
| Year published | 2022 |
| Volume and issue | 20(S2) |
| Article # | e200917 |
| DOI | 10.2903/j.efsa.2022.e200917 |
| ISSN | 1831-4732 |
| Distribution license | Creative Commons Attribution-NoDerivs 4.0 International |
| Website | https://efsa.onlinelibrary.wiley.com/doi/full/10.2903/j.efsa.2022.e200917 |
| Download | https://efsa.onlinelibrary.wiley.com/doi/pdfdirect/10.2903/j.efsa.2022.e200917 (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Risk and risk–benefit assessments of food are complex exercises, in which access to and use of several disconnected individual stand-alone databases is required to obtain hazard and exposure information. Data obtained from such databases ideally should be in line with the FAIR principles, i.e. the data must be findable, accessible, interoperable, and reusable. However, often cases are encountered when one or more of these principles are not followed. In this project, we set out to assess if existing commonly used databases in risk assessment are in line with the FAIR principles. We also investigated how access, interoperability, and reusability of data could be improved. We used the OpenFoodTox and the Seafood database as examples and showed how commonly used freely available open-source tools and repositories can be implemented in the data extraction process of risk assessments to increase data reusability and crosstalk across different databases.
Keywords: FAIR, food safety, risk assessment, OpenFoodTox, Seafood database, R, Shiny, Zenodo
Introduction
Description of work programme
Aims
This project assessed how to apply FAIR data principles [Wilkinson et al., 2016] in risk and risk–benefit assessments of food. Focusing on key databases recently used in a risk–benefit assessment of fish and seafood in the Norwegian diet [VKM et al., 2022a], the OpenFoodTox and the Seafood database, we aimed to demonstrate how open-source software tools can be used to make data stored in publicly available repositories more findable, accessible, interoperable, and reusable.
Methods
Using the [[R (programming language)|programming language R] [R Core Team, 2021] and data obtained from the European Food Safety Authority (EFSA) OpenFoodTox Tool [Dorne et al., 2020; Kovarich et al., 2022] and from the Institute of Marine Research (IMR) Seafood database [Institute of Marine Research, 2022], we assessed if programmatic optimization of access and the creation of a web-tool for selection and merging of subsets of the stored data improved findability, accessibility, interoperability, and reusability of the data. In this section, a brief description of both data and tools used is provided.
R
The programming language and environment R has been designed for the statistical analysis of data and the creation of graphics. [R Core Team, 2021] Over the past years, R has increasingly gained interest in the scientific research community [Hackenberger, 2020] as it is effective for data handling and includes many tools for basic and advanced data analysis. [R Core Team, 2021] R is a well-developed non-static language, which means that its base features can easily be extended via packages that can provide new functions and functionalities for different data science challenges, including bioinformatics and data mining. [Giorgi et al., 2022] In addition to this, R is supported by a big open-source community actively using this language and continuously adding new functionalities. R is licensed under the terms of the Free Software Foundation's GNU General Public License in source code form. [R Core Team, 2021] To facilitate programming with R, we used RStudio, an integrated development environment for R. [RStudio Team, 2022]
Shiny
As commented above, R can be expanded through packages, including one commonly used one called Shiny. [Chang et al., 2021] This package was designed with the idea of creating interactive web applications which use R in the backend. While the creator of a web-based Shiny-tool does need to know R, the end user of the web application created with Shiny does not need to have any knowledge of R. In addition to local installations of R and Shiny, Shiny web app also can be stored on a server, which users can access through their web browser. In both cases, the appearance and functionalities of the applications are the same, and the underlying R code can be shared freely.
Git and GitHub
Git is a version control system designed to allow different users to work on the same programming project, ensuring the traceability of progress and changes in the project. One of the most widely used providers of internet hosting for software development and version control using Git is GitHub. [Microsoft, 2022] GitHub implements Git and offers a free version, in which users can host different smaller projects and scripts, providing an easy way to share code created in R and other programming languages on the web. The scripts generated during this project will be hosted and accessible on GitHub in this repository. [Pineda-Pampliega, 2022]
Zenodo
Under the European OpenAIRE program, and with the idea of championing the sharing of scientific data, the Zenodo [European Organization for Nuclear Research, and OpenAIRE, 2013] open repository was developed and operated by CERN. [European Council for Nuclear Research] This open-source repository was developed for scientific data in a broad way, allowing to deposit not only research papers, but also data sets, software, reports, supplementary data and any other research-related digital artifacts. Submissions to Zenodo obtain a persistent digital object identifier (DOI), which facilitates the citation of the stored items and allows the sharing of data prior to their publication in peer-reviewed journals.
For a speedy exchange of evidence and supporting materials which could be used in food and feed safety risk assessments, EFSA has created a curated open repository called the Knowledge Junction within Zenodo. In addition to EFSA, several other institutions use Knowledge Junctions to share different data related to food security. For example, The Norwegian Scientific Committee for Food and Environment (VKM), which is part of this project, uses this Zenodo repository to upload finished reports (i.e., risk assessment and risk–benefit assessment) and supplementary materials of interest (i.e., literature searches, datasets, codes, etc.). To date, for VKM, the most recent example of the use of Zenodo is the opinion on the "Risk-Benefit Assessment of Sunscreen" [VKM et al., 2022b] For this opinion, the fellow Javier Pineda-Pampliega contributed to the preparation of the public sharing of the report's supplementary material, including datasets and R codes currently hosted on the VKM Knowledge Junction. [Norwegian Scientific Committee for Food and Environment (VKM), 2022]
Zenodo recently implemented the possibility to import GitHub workspaces; it now is possible to host completed GitHub projects also on Zenodo. This offers the advantage of obtaining a DOI for one's code, which simplifies the traceability and proper citation of code used to create the results.
OpenFoodTox
The EFSA's Chemical Hazards Database, OpenFoodTox [Dorne et al., 2020; Kovarich et al., 2022], is a structured database summarizing the outcomes of hazard identification and characterization for human and animal health and for the environment. It includes all regulated products and contaminants and provides open-source data for the (1) substance characterization, (2) links to EFSA outputs, and the values of (3) reference points, (4) reference values, and (5) genotoxicity. This database has become an essential tool for risk assessors and has provided the basis for the development and implementation of new approach methodologies (NAMs) in food and feed safety research. OpenFoodTox is hosted both on the EFSA webpage (as an interactive web tool) and on Zenodo in the EFSA Knowledge Junction.
Seafood database
The Institute of Marine Research in Norway routinely collects samples of key marine species for national and international monitoring programs. Their ISO/IEC 17025 accredited laboratories perform analyses of contaminants and nutrients using state-of-the-art methods. All the data generated, comprising multiple data points for over 25,000 individuals collected over a period of up to 15 years, are aggregated in a large in-house database. This database can be accessed freely through the online Seafood database portal [Institute of Marine Research, 2022], where the user can select between fish, shellfish, and seaweed divided by wild or farmed, and even prepared products, which can be found in Norwegian supermarkets. The database holds data of both Nutrients (separated into five categories: Amino acids, Fatty acids, Macro nutrients, Minerals, and Trace elements and Vitamins) and Contaminants (separated into four categories: Drug residues, Heavy metals, Organic pollutants, and Other undesirable substances).
Activities
With the aim to investigate the application of FAIR data principles in risk–benefit assessment of seafood, it was essential to evaluate opportunities and limitations in the OpenFoodTox and the Seafood database. Once evaluated, we developed publicly available R and Shiny code, which attempts to address potential limitations found and to add new functionalities for sub-setting and improved crosstalk between hazard and occurrence data repositories.
Evaluation and actions on the OpenFoodTox database
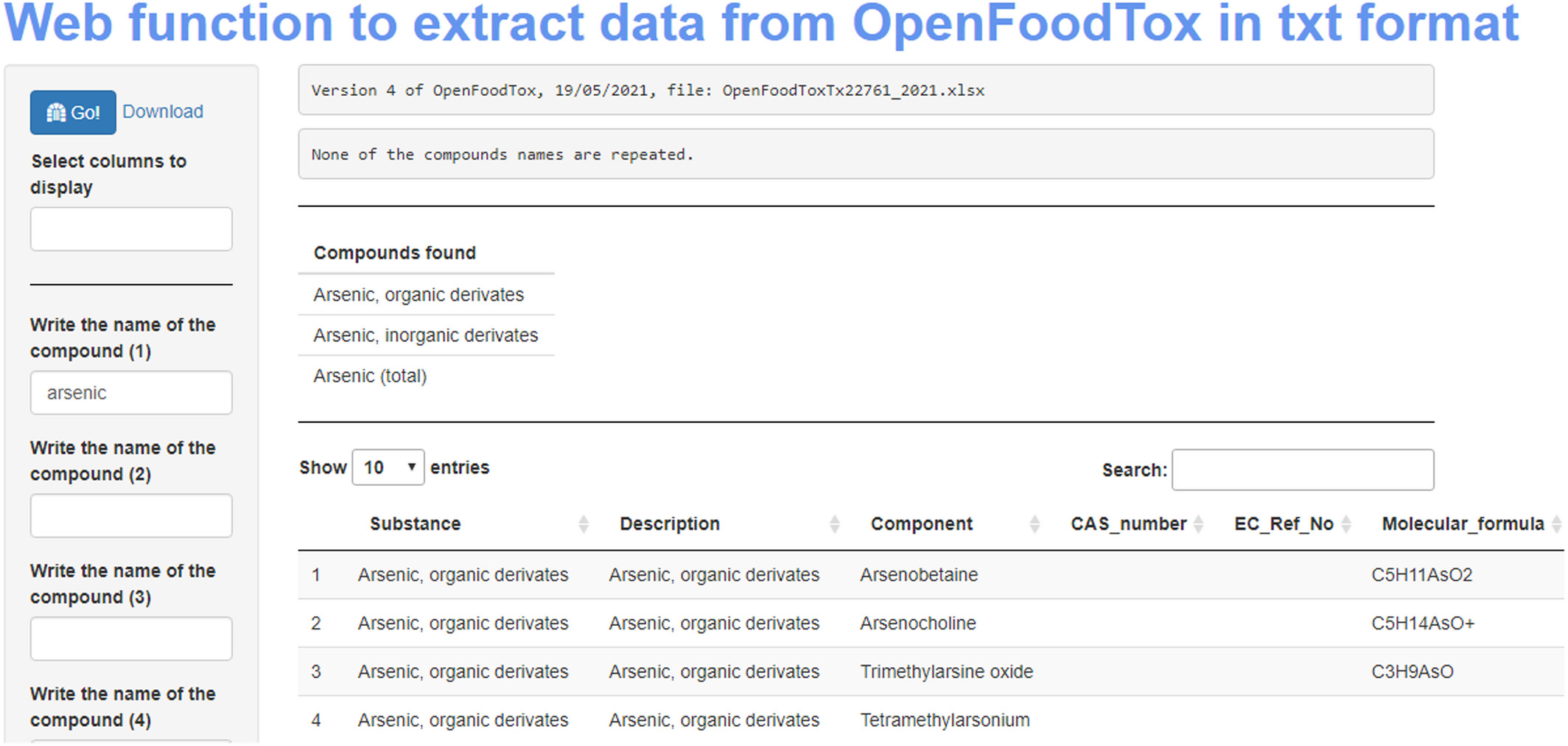
The OpenFoodTox database can be used in two different ways. The first (1) option is through the EFSA-hosted web application. The EFSA-hosted web application of the OpenFoodTox tool presents a classical interface, where different compounds can be searched by name. When searching, selected substances appear in five different categories of results: Substance characterization, EFSA outputs, Reference points, Reference values, and Genotoxicity. The resulting output represents the main limitation, as each category only can be downloaded individually (either in pdf, csv or xlsx format). In other words, after a search, the users need to download five different files and manually merge the data.
The second (2) option to access data is to download the entire OpenFoodTox database in xlsx format (Microsoft Excel Open XML Spreadsheet) from Zenodo. The data comprises five individual spreadsheets providing data on (1) substance characterization, (2) EFSA outputs, (3) reference points, (4) reference values, and (5) genotoxicity results. There is another “complete” spreadsheet, which is a combination of the five spreadsheets commented above (each one in a different tab) in addition to a dictionary spreadsheet. [Dorne et al., 2020] This makes data interoperable. However, as was described in the example above, to work with subsets of data spreading across the different spreadsheets, data aggregation and merging again must be performed manually using additional software for tabular data files. The most common among these tools is Excel, which is part of the commercial Microsoft Office Suite, but other free alternatives such as OpenOffice, LibreOffice, or online tools such as Google Drive Sheets also can be used. In any case, for merging the large individual datasets, the user needs to be proficient in the terminology of terms and use of spreadsheet tools for efficient filtering, merging, and sub-setting of the data in the desired format.
To evaluate potential complementary solutions to access, subset, and merge data stored in the EFSA OpenFoodTox database on Zenodo, in the present project using R (vers. 4.1.2) running in RStudio (vers. 2022.2.3.492), functions (i.e., pieces of code which work together for a common purpose) were written using R markdown, being characterized by the following features:
- Data can be downloaded directly from the OpenFoodTox URL to eliminate the need for the user to search for and/or download the data in Excel.
- The database offers the possibility to search for up to 15 elements at the same time, with an implemented control of any repeated entry values. In the case of repetition, the repeated value is indicated, but not considered in the search.
- If a search is entered for a general term and several compounds appear in the database, an indication for the number of the different compounds is provided. For example, the search “lead” returns four results, because the components identified in the database are: “Lead,” “Lead (II),” “Lead sulphate,” and “Tetraethyl lead.”
- To increase the (computational) reusability of the data in automated analysis pipelines, the information is downloaded in a plain text file (txt). This is a standard format of plain text that can be open in many different software tools. However, also the possibility to download data in csv (comma-separated values) is provided.
- To increase traceability information on the OpenFoodTox database version and the date and time when the file was created are automatically appended to the name of the downloaded file.
After the creation of the R script, to increase the number of potential users of this tool, we assessed if an additional approach that does not require knowledge and use of R could be developed. For this, the creation of a web-based application using Shiny was attempted. The use of Shiny opens the possibility to access and subset OpenFoodTox data using an internet browser only and also allows for the implementation of additional functions into our R code. That is, in addition to the characteristics of the function described above, the Shiny application developed in this project (Figure 1) has the following extra functions:
- Increased traceability: an indication of which version of the OpenFoodTox database used has been included. At the time of writing this report, the fifth iteration of the OpenFoodTox was released (and published on Zenodo on 16 June 2022).
- Implementation of interactive tables, allowing to filter results in real-time.
- Initially, tables will show all columns in the dataset, but tools for sub-setting and selection of individual columns to be retained are provided. This functionality makes it easier to take snapshots only of the columns of interest for further uses.
- With one of the objectives of this project being to facilitate the interaction and crosstalk between databases of interest to risk assessors, the option to add links to PubChem for each selected compound was implemented. PubChem is a database of chemical molecules and their activities, maintained by the National Centre for Biotechnology Information (NCBI) of the United States.
|
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation and updates to spelling and grammar. In some cases important information was missing from the references, and that information was added. No other changes have been made, in accord with the "NoDerivs" portion of the license.