Journal:iLAP: A workflow-driven software for experimental protocol development, data acquisition and analysis
| Full article title | iLAP: a workflow-driven software for experimental protocol development, data acquisition and analysis |
|---|---|
| Journal | BMC Bioinformatics |
| Author(s) | Stocker, Gernot; Fischer, Maria; Rieder, Dietmar; Bindea, Gabriela; Kainz, Simon; Oberstolz, Michael; McNally, James G.; Trajanoski, Zlatko |
| Author affiliation(s) | Institute for Genomics and Bioinformatics, Graz University of Technology; National Cancer Institute, National Institutes of Health |
| Primary contact | Email: zlatko.trajanoski@tugraz.at |
| Year published | 2009 |
| Volume and issue | 10 |
| Page(s) | 390 |
| DOI | 10.1186/1471-2105-10-390 |
| ISSN | 1471-2105 |
| Distribution license | Creative Commons Attribution 2.0 |
| Website | http://www.biomedcentral.com/1471-2105/10/390 |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Background
In recent years, the genome biology community has expended considerable effort to confront the challenges of managing heterogeneous data in a structured and organized way and developed laboratory information management systems (LIMS) for both raw and processed data. On the other hand, electronic notebooks were developed to record and manage scientific data, and facilitate data-sharing. Software which enables both, management of large datasets and digital recording of laboratory procedures would serve a real need in laboratories using medium and high-throughput techniques.
Results
We have developed iLAP (Laboratory data management, Analysis, and Protocol development), a workflow-driven information management system specifically designed to create and manage experimental protocols, and to analyze and share laboratory data. The system combines experimental protocol development, wizard-based data acquisition, and high-throughput data analysis into a single, integrated system. We demonstrate the power and the flexibility of the platform using a microscopy case study based on a combinatorial multiple fluorescence in situ hybridization (m-FISH) protocol and 3D-image reconstruction. iLAP is freely available under the open source license AGPL from http://genome.tugraz.at/iLAP/. (Webcite)
Conclusion
iLAP is a flexible and versatile information management system, which has the potential to close the gap between electronic notebooks and LIMS and can therefore be of great value for a broad scientific community.
Background
The development of novel large-scale technologies has considerably changed the way biologists perform experiments. Genome biology experiments do not only generate a wealth of data, but they often rely on sophisticated laboratory protocols comprising hundreds of individual steps. For example, the protocol for chromatin immunoprecipitation on a microarray (Chip-chip) has 90 steps, uses over 30 reagents and 10 different devices.[1] Even adopting an established protocol for large-scale studies represents a daunting challenge for the majority of the labs. The development of novel laboratory protocols and/or the optimization of existing ones is still more distressing, since this requires systematic changes of many parameters, conditions, and reagents. Such changes are becoming increasingly difficult to trace using paper lab books. A further complication for most protocols is that many laboratory instruments are used, which generate electronic data stored in an unstructured way at disparate locations. Therefore, protocol data files are seldom or never linked to notes in lab books and can be barely shared within or across labs. Finally, once the experimental large-scale data have been generated, they must be analyzed using various software tools, then stored and made available for other users. Thus, it is apparent that software support for current biological research — be it genomic or performed in a more traditional way — is urgently needed and inevitable.
In recent years, the genome biology community has expended considerable effort to confront the challenges of managing heterogeneous data in a structured and organized way and as a result developed information management systems for both raw and processed data. Laboratory information management systems (LIMS) have been implemented for handling data entry from robotic systems and tracking samples[2][3] as well as data management systems for processed data including microarrays[4][5], proteomics data[6][7][8], and microscopy data.[9] The latter systems support community standards like FUGE[10][11], MIAME[12], MIAPE[13], or MISFISHIE[14] and have proven invaluable in a state-of-the-art laboratory. In general, these sophisticated systems are able to manage and analyze data generated for only a single type or a limited number of instruments, and were designed for only a specific type of molecule.
On the other hand, commercial as well as open source electronic notebooks[15][16][17][18][19] were developed to record and manage scientific data, and facilitate data-sharing. The influences encouraging the use of electronic notebooks are twofold.[16] First, much of the data that needs to be recorded in a laboratory notebook is generated electronically. Transcribing data manually into a paper notebook is error-prone, and in many cases, for example, analytical data (spectra, chromatograms, photographs, etc.), transcription of the data is not possible. Second, the incorporation of high-throughput technologies into the research process has resulted in an increased volume of electronic data that need to be transcribed. As opposed to LIMS, which captures highly structured data through rigid user interfaces with standard report formats, electronic notebooks contain unstructured data and have flexible user interfaces.
Software which enables both, management of large datasets and recording of laboratory procedures, would serve a real need in laboratories using medium and high-throughput techniques. To the best of our knowledge, there is no software system available, which supports tedious protocol development in an intuitive way, links the plethora of generated files to the appropriate laboratory steps and integrates further analysis tools. We have therefore developed iLAP, a workflow-driven information management system for protocol development and data management. The system combines experimental protocol development, wizard-based data acquisition, and high-throughput data analysis into a single, integrated system. We demonstrate the power and the flexibility of the platform using a microscopy case study based on combinatorial multiple fluorescence in situ hybridization (m-FISH) protocol and 3D-image reconstruction.
Implementation
Workflow-driven software design
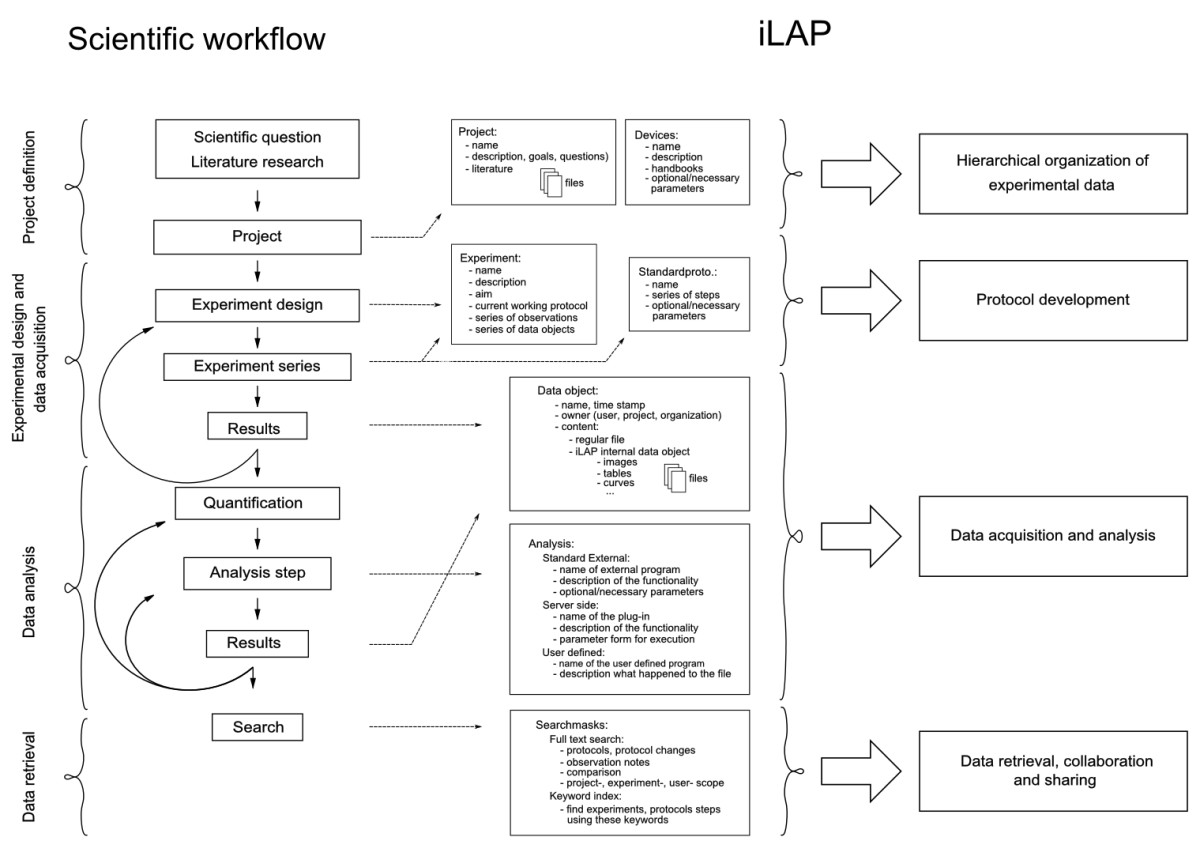
The design of a software platform that supports the development of protocols and data management in an experimental context has to be based on and directed by the laboratory workflow. The laboratory workflow can be divided into four principal steps: 1) project definition phase, 2) experimental design and data acquisition phase, 3) data analysis and processing phase and 4) data retrieval phase (Figure 1).
Figure 1: Mapping of the laboratory workflow onto iLAP features. The software design of iLAP is inspired by a typical laboratory workflow in life sciences and offers software assistance during the process.
The figure illustrates on the left panel the scientific workflow separated into four phases: project definition, data acquisition and analysis, and data retrieval. The right panel shows the main functionalities offered by iLAP.
Project definition phase
A scientific project starts with a hypothesis and the choice of methods required to address a specific biological question. Already during this initial phase it is crucial to define the question as specifically as possible and to capture the information in a digital form. Documents collected during the literature research should be collated with the evolving project definition for later review or for sharing with other researchers. All files collected in this period should be attached to the defined projects and experiments in the software.
Experimental design and data acquisition
Following the establishment of a hypothesis and based on preliminary experiments, the detailed design of the biological experiments is then initiated. Usually, the experimental work follows already established standard operating procedures, which have to be modified and optimized for the specific biological experiment. These protocols are defined as a sequence of protocol steps. However, well-established protocols must be kept flexible in a way that particular conditions can be changed. The typically changing parameters of standard protocol steps (e.g. fixation times, temperature changes etc.) are important to record as they are used to improve the experimental reproducibility.
Equipped with a collection of standard operating procedures, an experiment can be initiated and the data generated. In general, data acquisition comprises not only files but also observations of interest, which might be relevant for the interpretation of the results. Most often these observations disappear in paper notebooks and are not accessible in a digital form. Hence, these experimental notes should be stored and attached to the originating protocol step, experiment or project.
Data analysis and processing
After storing the raw result files, additional analysis and post-processing steps must be performed to obtain processed data for subsequent analysis. In order to extract information and to combine it in a statistically meaningful manner, multiple data sets have to be acquired. The software workflow should enable also the inclusion of external analytical steps, so that files resulting from external analysis software can be assigned to their original raw data files. Finally, the data files generated at the analysis stage should be connected to the raw data, allowing connection of the data files with the originating experimental context.
Data retrieval
By following the experimental workflow, all experimental data e.g. different files, protocols, notes etc. should be organized in a chronological and project-oriented way and continuously registered during their acquisition. An additional advantage should be the ability to search and retrieve the data. Researchers frequently have to search through notebooks to find previously uninterpretable observations. Subsequently, as the project develops, the researchers gain a different perspective and recognize that prior observations could lead to new discoveries. Therefore, the software should offer easy to use interfaces that allow searches through observation notes, projects- and experiment descriptions.
Software architecture
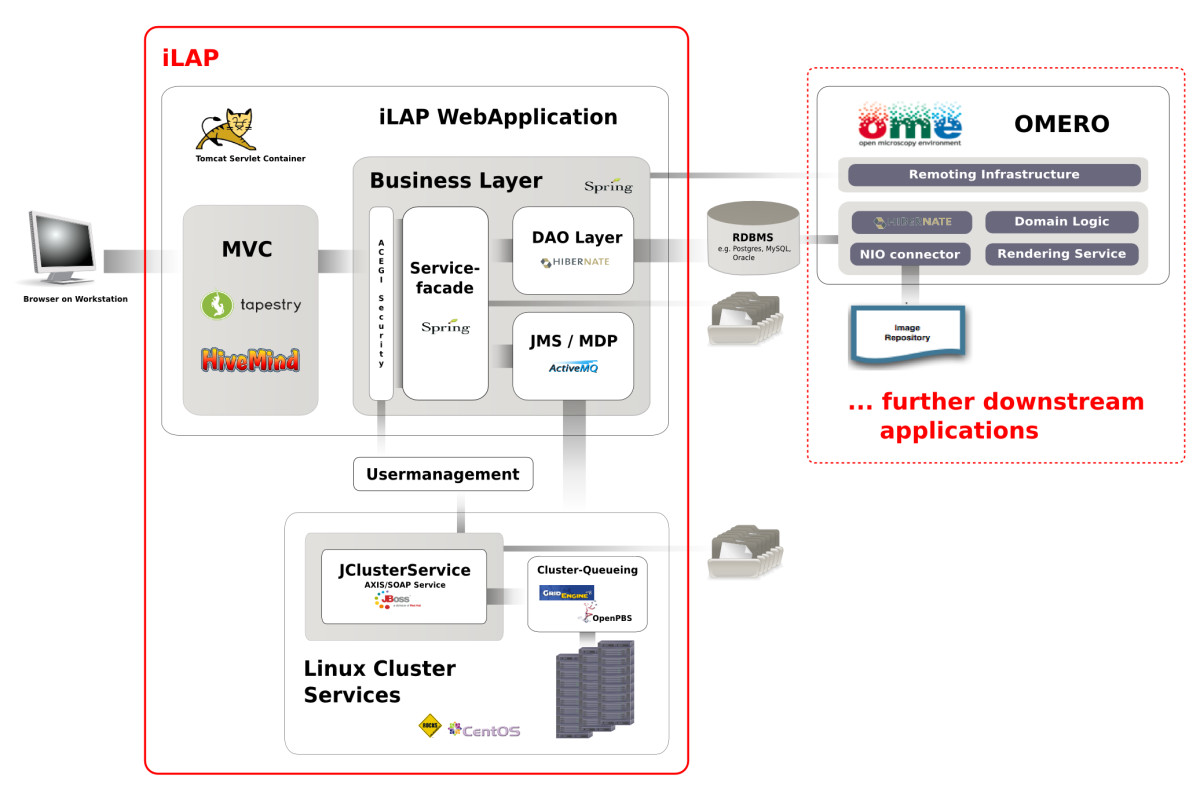
iLAP is a multi-tier client-server application and can be subdivided into different functional modules which interact as self-contained units according to their defined responsibilities (see Figure 2).
Figure 2: Software Architecture. iLAP features a typical three-tier architecture and can hence be divided into a presentation tier, business tier and a persistence tier (from left to right). The presentation tier is formed by a graphical user interface, accessed using a web browser. The following business layer is protected by a security layer, which enforces user authentication and authorization. After access is granted, the security layer passes the user requests to the business layer, which is mainly responsible for guiding the user through the laboratory workflow. This layer also coordinates all background tasks like automatic surveying of analysis jobs on a computing cluster or synchronizing/exchanging data with further downstream applications. (e.g. OMERO (open microscopy environment) image server). Finally, the persistence layer interacts with the relational database.
Presentation tier
The presentation tier within iLAP is formed by a Web interface, using Tapestry[20] as the model view controller and an Axis Web service[21], which allows programming access to parts of the application logic. Thus, on the client side, a user requires an Internet connection and a recent Web browser with Java Applet support, available for almost every platform. In order to provide a simple, consistent but also attractive Web interface, iLAP follows usability guidelines described in[22][23] and uses Web 2.0 technologies for dynamic content generation.
Business tier and runtime environment
The business tier is realized as view-independent application logic, which stores and retrieves datasets by communicating with the persistence layer. The internal management of files is also handled from a central service component, which persists the meta-information for acquired files to the database, and stores the file content in a file-system-based data hierarchy. The business layer also holds asynchronous services for application-internal JMS messaging and for integration of external computing resources like high-performance computing clusters. All services of this layer are implemented as Spring[24] beans, for which the Spring-internal interceptor classes provide transactional integrity.
The business tier and the persistence tier are bound by the Spring J2EE lightweight container, which manages the component-object life cycle. Furthermore, the Spring context is transparently integrated into the Servlet context of Tapestry using the HiveMind[25] container backend. This is realized by using the automatic dependency injection functionality of HiveMind which avoids integrative glue code for lookups into the Spring container. Since iLAP uses Spring instead of EJB related components, the deployment of the application only requires a standard conformed Servlet container. Therefore, the Servlet container Tomcat[26] is used, which offers not only Servlet functionality but J2EE infrastructure services[27] such as centrally configured data-sources and transaction management realized with the open source library JOTM.[28] This makes the deployment of iLAP on different servers easier, because machine-specific settings for different production environments are kept outside the application configuration.
External programming interfaces
The SOAP Web service interface for external programmatic access is realized by combining the Web service framework Axis with corresponding iLAP components. The Web service operates as an external access point for Java Applets within the Web application, as well as for external analysis and processing applications such as ImageJ. Model driven development
In order to reduce coding and to increase the long term maintainability, the model driven development environment AndroMDA[29] is used to generate components of the persistence layer and recurrent parts from the above mentioned business layer. AndroMDA accomplishes this by translating an annotated UML-model into a JEE-platform-specific implementation using Hibernate and Spring as base technology. Due to the flexibility of AndroMDA, application external services, such as the user management system, have a clean integration in the model. Dependencies of internal service components on such externally defined services are cleanly managed by its build system.
By changing the build parameters in the AndroMDA configuration, it is also possible to support different relational database management systems. This is because platform specific code with the same functionality is generated for data retrieval. Furthermore, technology lock-in regarding the implementation of the service layers was also addressed by using AndroMDA, as the implementation of the service facade can be switched during the build process from Spring based components to distributed Enterprise Java Beans. At present, iLAP is operating on one local machine and, providing the usage scenarios do not demand it, this architectural configuration will remain. However, chosen technologies are known to work on Web server farms and crucial distribution of the application among server nodes is transparently performed by the chosen technologies.
Asynchronous data processing
The asynchronous handling of business processes is realized in iLAP with message-driven Plain Old Java Objects (POJOs). Hence, application tasks, such as the generation of image previews, can be performed asynchronously. If performed immediately, these would unnecessarily block the responsiveness of the Web front-end. iLAP delegates tasks via JMS messages to back-end services, which perform the necessary processing actions in the background.
These back-end services are also UML-modelled components and receive messages handled by the JMS provider ActiveMQ. If back-end tasks consume too many calculation resources, the separation of Web front-end and JMS message receiving services can be realized by copying the applications onto two different servers and changing the Spring JMS configuration.
For the smooth integration of external computing resources like the high-performance computing cluster or special compute nodes with limited software licenses the JClusterService is used. JClusterService is a separately developed J2EE application which enables a programmer to run generic applications on a remote execution host or high-performance computing cluster. Every application which offers a command line interface can be easily integrated by defining a service definition in XML format and accessing it via a SOAP-based programming interface from any Java-application. The execution of the integrated application is carried out either by using the internal JMS-queuing system for single host installations or by using the open source queuing systems like Sun Grid Engine (Sun Microsystems) or OpenPBS/Torque.
Results
Functional overview
The functionality offered by the iLAP web interface can be described by four components: 1) hierarchical organization of the experimental data, 2) protocol development, 3) data acquisition and analysis, and 4) data retrieval and data sharing (Figure 1). iLAP specific terms are summarized in Table 1.
| ||||||||||||||||||||||||||||
References
- ↑ Acevedo, L.G.; Iniguez, A.L.; Holster, H.L.; Zhang, X.; Green, R.; Farnham, P.J. (2007). "Genome-scale ChIP-chip analysis using 10,000 human cells". Biotechniques 43 (6): 791-797. PMC PMC2268896. PMID 18251256. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2268896.
- ↑ Piggee, C. (2008). "LIMS and the art of MS proteomics". Analytical Chemistry 80 (13): 4801-4806. doi:10.1021/ac0861329. PMID 18609747.
- ↑ Haquin, S.; Oeuillet, E.; Pajon, A.; Harris, M.; Jones, A.T.; van Tilbeurgh, H.; et al. (2008). "Data management in structural genomics: an overview". Methods in Molecular Biology 426: 49-79. doi:10.1007/978-1-60327-058-8_4. PMID 18542857.
- ↑ Maurer, M.; Molidor, R.; Sturn, A.; Hartler, J.; Hackl, H.; Stocker, G.; et al. (2005). "MARS: microarray analysis, retrieval, and storage system". BMC Bioinformatics 6: 101. doi:10.1186/1471-2105-6-101. PMC PMC1090551. PMID 15836795. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1090551.
- ↑ Saal, L.H.; Troein, C.; Vallon-Christersson, J.; Gruvberger, S.; Borg, A.; Peterson, C. (2002). "BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data". Genome Biology 3 (8): SOFTWARE0003. doi:10.1186/gb-2002-3-8-software0003. PMC PMC139402. PMID 12186655. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC139402.
- ↑ Hartler, J.; Thallinger, G.G.; Stocker, G.; Sturn, A.; Burkard, T.R.; Korner, E.; et al. (2007). "MASPECTRAS: a platform for management and analysis of proteomics LC-MS/MS data". BMC Bioinformatics 8: 197. doi:10.1186/1471-2105-8-197. PMC PMC1906842. PMID 17567892. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1906842.
- ↑ Craig, R.; Cortens, J.P.; Beavis, R.C. (2004). "Open source system for analyzing, validating, and storing protein identification data". Journal of Proteome Research 3 (6): 1234-1242. doi:10.1021/pr049882h. PMID 15595733.
- ↑ Rauch, A.; Bellew, M.; Eng, J.; Fitzgibbon, M.; Holzman, T.; Hussey, P.; et al. (2006). "Computational Proteomics Analysis System (CPAS): an extensible, open-source analytic system for evaluating and publishing proteomic data and high throughput biological experiments". Journal of Proteome Research 5 (1): 112-121. doi:10.1021/pr0503533. PMID 16396501.
- ↑ Moore, J.; Allan, C.; Burel, J.M.; Loranger, B.; MacDonald, D.; Monk, J.; et al. (2008). "Open tools for storage and management of quantitative image data". Methods in Cell Biology 85: 555-570. doi:10.1016/S0091-679X(08)85024-8. PMID 18155479.
- ↑ Jones, A.R.; Pizarro, A.; Spellman, P.; Miller, M. (2006). "FuGE: Functional Genomics Experiment Object Model". OMICS 10 (2): 179-184. doi:10.1089/omi.2006.10.179. PMID 16901224.
- ↑ Jones, A.R.; Miller, M.; Aebersold, R.; Apweiler, R.; Ball, C.A.; Brazma, A.; et al. (2007). "The Functional Genomics Experiment model (FuGE): an extensible framework for standards in functional genomics". Nature Biotechnology 25 (10): 1127-1133. doi:10.1038/nbt1347. PMID 17921998.
- ↑ Brazma, A.; Hingamp, P.; Quackenbush, J.; Sherlock, G.; Spellman, P.; Stoeckert, C.; et al. (2001). "Minimum information about a microarray experiment (MIAME)-toward standards for microarray data". Nature Genetics 29 (4): 365-371. doi:10.1038/ng1201-365. PMID 11726920.
- ↑ Taylor, C.F.; Paton, N.W.; Lilley, K.S.; Binz, P.A.; Julian, Jr., R.K.; Jones AR, et al. (2007). "The minimum information about a proteomics experiment (MIAPE)". Nature Biotechnology 25 (8): 887-893. doi:10.1038/nbt1329. PMID 17687369.
- ↑ Deutsch, E.W.; Ball, C.A.; Berman, J.J.; Bova, G.S.; Brazma, A.; Bumgarner, R.E.; et al. (2008). "Minimum information specification for in situ hybridization and immunohistochemistry experiments (MISFISHIE)". Nature Biotechnology 26 (3): 305-312. doi:10.1038/nbt1391. PMC PMC4367930. PMID 18327244. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4367930.
- ↑ Taylor, K.T. (2007). "The status of electronic laboratory notebooks for chemistry and biology". Current Opinion in Drug Discovery & Development 12 (15-16): 647-649. doi:10.1016/j.drudis.2007.06.010. PMID 17706546.
- ↑ 16.0 16.1 Drake, D.J. (2006). "ELN implementation challenges". Drug Discovery Today 9 (3): 348-353. PMID 16729731.
- ↑ Butler, D. (2005). "Electronic notebooks: a new leaf". Nature 436 (7047): 20-21. doi:10.1038/436020a. PMID 16001034.
- ↑ Kihlen, M. (2005). "Electronic lab notebooks - do they work in reality?". Drug Discovery Today 10 (18): 1205-1207. doi:10.1016/S1359-6446(05)03576-2. PMID 16213407.
- ↑ Bradley, J.-C.; Samuel, B. (2004). "SMIRP-A Systems Approach to Laboratory Automation". Journal of the Association for Laboratory Automation 5 (3): 48-53. doi:10.1016/S1535-5535(04)00074-7.
- ↑ "Tapestry web frame work". Apache Software Foundation. 2009. Archived from the original on 23 July 2010. http://www.webcitation.org/query.php?url=http://tapestry.apache.org/&refdoi=10.1186/1471-2105-10-390.
- ↑ "Java implementation of the SOAP ("Simple Object Access Protocol")". Apache Software Foundation. 2009. Archived from the original on 07 December 2005. http://www.webcitation.org/query.php?url=http://ws.apache.org/axis/&refdoi=10.1186/1471-2105-10-390.
- ↑ Krug, S. (2000). Don't make me think! A Common Sense Apporach to Web Usability. Indianapolis, IN: New Riders Publishing.
- ↑ Johnson, J. (2003). Web Bloopers: 60 Common Web Design Mistakes and How to Avoid Them. San Francisco, CA: Morgan Kaufmann Publishers, Inc.
- ↑ "Spring lightweight application container". SpringSource. 2009. Archived from the original on 27 May 2009. http://www.webcitation.org/query.php?url=http://www.springsource.org/&refdoi=10.1186/1471-2105-10-390.
- ↑ "Services and configuration microkernel". Apache Software Foundation. 2009. http://hivemind.apache.org/.
- ↑ "Apache servlet container". Apache Software Foundation. 2009. Archived from the original on 27 May 2009. http://www.webcitation.org/query.php?url=http://tomcat.apache.org/&refdoi=10.1186/1471-2105-10-390.
- ↑ Johnson, R.; Hoeller, J. (2004). Expert One-on-One J2EE Development without EJB. Wrox.
- ↑ "Java Open Transaction Manager (JOTM)". OW2 Consortium. 2009. http://jotm.ow2.org/xwiki/bin/view/Main/WebHome?.
- ↑ Bohlen, M. (2009). "AndroMDA". Archived from the original on 27 March 2007. http://www.webcitation.org/query.php?url=http://www.andromda.org/&refdoi=10.1186/1471-2105-10-390.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In most of the article's references DOIs and PubMed IDs were not given; they've been added to make the references more useful.